You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002481_00238
You are here: Home > Sequence: MGYG000002481_00238
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Stenotrophomonas maltophilia_F | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Stenotrophomonas; Stenotrophomonas maltophilia_F | |||||||||||
| CAZyme ID | MGYG000002481_00238 | |||||||||||
| CAZy Family | GH31 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 127003; End: 130362 Strand: - | |||||||||||
Full Sequence Download help
| MENCVRRSPL RLALVPALML ASTTARAEPV GNLRTVSASA SRDGVQGWDL QTDKGARIRI | 60 |
| ELPATDIIRV QAGRNGTLTG AGDKAAPIVL PQPKASVQAQ LEEDAQEIRV RTEALVLHVQ | 120 |
| RQPLRLRLER LDNGQPTALW QELQPLDLDA TQSVQVLSSQ ADEGYFGGGQ QNGRYQFKGR | 180 |
| ELEVSYSGGW EEGDRPSPAP MLLSSRGWGM LRNTWSDGSY DLRQSDQATL LHREDRFDAY | 240 |
| YFVGADLPKL IERYTQLTGR PNMVARWALS YGDADCYNDG DNSRKPGTVP EGWSDGPTGT | 300 |
| TPDVIDSVAK QYRAHDMPGG WILPNDGYGC GYKQLPETVK GLAGYGFRTG LWTENGVDKI | 360 |
| AWEVGKAGSR VQKLDVAWTG KGYQFAMDAN RQAFNGILDN SDSRPFLWTV MGWAGIQRYA | 420 |
| VAWTGDQSSS WDYIRWHVPT LVGSGLSGMA YASGDVDAIF GGSAETFTRD LQWKAFTPVL | 480 |
| MGMSGWSSNA RKHPWWYDEP YRSINRDYLK LKMRLTPYMY GLVHEAAQTG APPVRGLMWD | 540 |
| NPRDPHAQDE TYKYQFLLGR DLLVAPVHRS QAASRGWRRD IHLPAGGWID YWDGRRVQAA | 600 |
| ADGRQLDRQV DLATLPVFVR AGAILPMYPS MLFDGEKPLD EVTFDLYPQG DSQYTLYEDD | 660 |
| GNTRRYQQGE SSTQVIRVQA PAQGSGPVQV QIDAVQGQYN GQLAQRRYGL RVLSRQAPRA | 720 |
| VQAGGRALPA LADAAAFNSA NEGWYFDAKD RRGTVHVRTA TQDIRQPLQL QLDFAVAAAA | 780 |
| ADDAYPAAPV LGRELPADSL LVVNRPAEEP GHALENAFDD DPATWFRSVR NQAVRTGAHE | 840 |
| WVIGFGERKM IDGIDIAPRN DKNWKHGQVR DYEVYLGDSN GEWGEPIARG RLQLKEGVQR | 900 |
| IDFPAHAGRL LRFRVMSVQN PEGDGASSTD PMVTAAQGSA RAFDALQPRD VGPIALSTFH | 960 |
| ILEHQEPERP ARQRYLSELP VPAALASQVR TDQSFRGDAG MRMNGLQFRR GLGVGASSRI | 1020 |
| DLRLQGGWHL LRADLGIDDA CRSAGGLQFQ VWGDNRLLYD SGLVKAPGVV KPELDIRGLS | 1080 |
| TLSLRTLGAQ GSQPAQVCAN WANAVLIGQE GDSASIVAP | 1119 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH31 | 386 | 625 | 6.6e-65 | 0.5386416861826698 |
| CBM51 | 975 | 1106 | 2.9e-27 | 0.9850746268656716 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06596 | GH31_CPE1046 | 0.0 | 257 | 594 | 1 | 334 | Clostridium CPE1046-like. CPE1046 is an uncharacterized Clostridium perfringens protein with a glycosyl hydrolase family 31 (GH31) domain. The domain architecture of CPE1046 and its orthologs includes a C-terminal fibronectin type 3 (FN3) domain and a coagulation factor 5/8 type C domain in addition to the GH31 domain. Enzymes of the GH31 family possess a wide range of different hydrolytic activities including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-xylosidase, 6-alpha-glucosyltransferase, 3-alpha-isomaltosyltransferase and alpha-1,4-glucan lyase. All GH31 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. |
| COG1501 | YicI | 3.22e-75 | 58 | 691 | 48 | 737 | Alpha-glucosidase, glycosyl hydrolase family GH31 [Carbohydrate transport and metabolism]. |
| pfam01055 | Glyco_hydro_31 | 2.36e-61 | 240 | 625 | 1 | 442 | Glycosyl hydrolases family 31. Glycosyl hydrolases are key enzymes of carbohydrate metabolism. Family 31 comprises of enzymes that are, or similar to, alpha- galactosidases. |
| cd06603 | GH31_GANC_GANAB_alpha | 8.17e-44 | 394 | 663 | 201 | 467 | neutral alpha-glucosidase C, neutral alpha-glucosidase AB. This subgroup includes the closely related glycosyl hydrolase family 31 (GH31) isozymes, neutral alpha-glucosidase C (GANC) and the alpha subunit of heterodimeric neutral alpha-glucosidase AB (GANAB). Initially distinguished on the basis of differences in electrophoretic mobility in starch gel, GANC and GANAB have been shown to have other differences, including those of substrate specificity. GANC and GANAB are key enzymes in glycogen metabolism that hydrolyze terminal, non-reducing 1,4-linked alpha-D-glucose residues from glycogen in the endoplasmic reticulum. The GANC/GANAB family includes the alpha-glucosidase II (ModA) from Dictyostelium discoideum as well as the alpha-glucosidase II (GLS2, or ROT2 - Reversal of TOR2 lethality protein 2) from Saccharomyces cerevisiae. |
| cd06604 | GH31_glucosidase_II_MalA | 3.33e-30 | 385 | 530 | 193 | 339 | Alpha-glucosidase II-like. Alpha-glucosidase II (alpha-D-glucoside glucohydrolase) is a glycosyl hydrolase family 31 (GH31) enzyme, found in bacteria and plants, which has exo-alpha-1,4-glucosidase and oligo-1,6-glucosidase activities. Alpha-glucosidase II has been characterized in Bacillus thermoamyloliquefaciens where it forms a homohexamer. This subgroup also includes the MalA alpha-glucosidase from Sulfolobus solfataricus and the AglA alpha-glucosidase from Picrophilus torridus. MalA is part of the carbohydrate-metabolizing machinery that allows this organism to utilize carbohydrates, such as maltose, as the sole carbon and energy source. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWB80361.1 | 0.0 | 1 | 1119 | 1 | 1119 |
| CCH14506.1 | 0.0 | 1 | 1119 | 1 | 1119 |
| QCB34207.1 | 0.0 | 1 | 1119 | 1 | 1119 |
| QGL86366.1 | 0.0 | 1 | 1119 | 1 | 1119 |
| VEF35171.1 | 0.0 | 1 | 1119 | 1 | 1119 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6M76_A | 1.36e-104 | 131 | 723 | 141 | 787 | GH31alpha-N-acetylgalactosaminidase from Enterococcus faecalis [Enterococcus faecalis ATCC 10100],6M77_A GH31 alpha-N-acetylgalactosaminidase from Enterococcus faecalis in complex with N-acetylgalactosamine [Enterococcus faecalis ATCC 10100] |
| 7F7R_A | 6.82e-104 | 131 | 723 | 141 | 787 | ChainA, GH31 alpha-N-acetylgalactosaminidase [Enterococcus faecalis ATCC 10100] |
| 7F7Q_A | 1.80e-103 | 131 | 723 | 141 | 787 | ChainA, GH31 alpha-N-acetylgalactosaminidase [Enterococcus faecalis ATCC 10100] |
| 6JR6_A | 8.71e-41 | 162 | 728 | 155 | 791 | Flavobacteriumjohnsoniae GH31 dextranase, FjDex31A [Flavobacterium johnsoniae UW101],6JR6_B Flavobacterium johnsoniae GH31 dextranase, FjDex31A [Flavobacterium johnsoniae UW101],6JR6_C Flavobacterium johnsoniae GH31 dextranase, FjDex31A [Flavobacterium johnsoniae UW101],6JR6_D Flavobacterium johnsoniae GH31 dextranase, FjDex31A [Flavobacterium johnsoniae UW101],6JR7_A Flavobacterium johnsoniae GH31 dextranase, FjDex31A, complexed with glucose [Flavobacterium johnsoniae UW101],6JR7_B Flavobacterium johnsoniae GH31 dextranase, FjDex31A, complexed with glucose [Flavobacterium johnsoniae UW101],6JR7_C Flavobacterium johnsoniae GH31 dextranase, FjDex31A, complexed with glucose [Flavobacterium johnsoniae UW101],6JR7_D Flavobacterium johnsoniae GH31 dextranase, FjDex31A, complexed with glucose [Flavobacterium johnsoniae UW101] |
| 6JR8_A | 2.03e-40 | 162 | 728 | 155 | 791 | Flavobacteriumjohnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101],6JR8_B Flavobacterium johnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101],6JR8_C Flavobacterium johnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101],6JR8_D Flavobacterium johnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9P999 | 4.13e-39 | 200 | 670 | 153 | 667 | Alpha-xylosidase OS=Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) OX=273057 GN=xylS PE=1 SV=1 |
| Q9F234 | 6.06e-39 | 383 | 679 | 442 | 729 | Alpha-glucosidase 2 OS=Bacillus thermoamyloliquefaciens OX=1425 PE=3 SV=1 |
| B3PEE6 | 5.92e-28 | 47 | 661 | 47 | 711 | Oligosaccharide 4-alpha-D-glucosyltransferase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=agd31B PE=1 SV=1 |
| P79403 | 4.64e-26 | 404 | 669 | 596 | 857 | Neutral alpha-glucosidase AB OS=Sus scrofa OX=9823 GN=GANAB PE=1 SV=1 |
| Q14697 | 1.39e-25 | 388 | 667 | 585 | 855 | Neutral alpha-glucosidase AB OS=Homo sapiens OX=9606 GN=GANAB PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
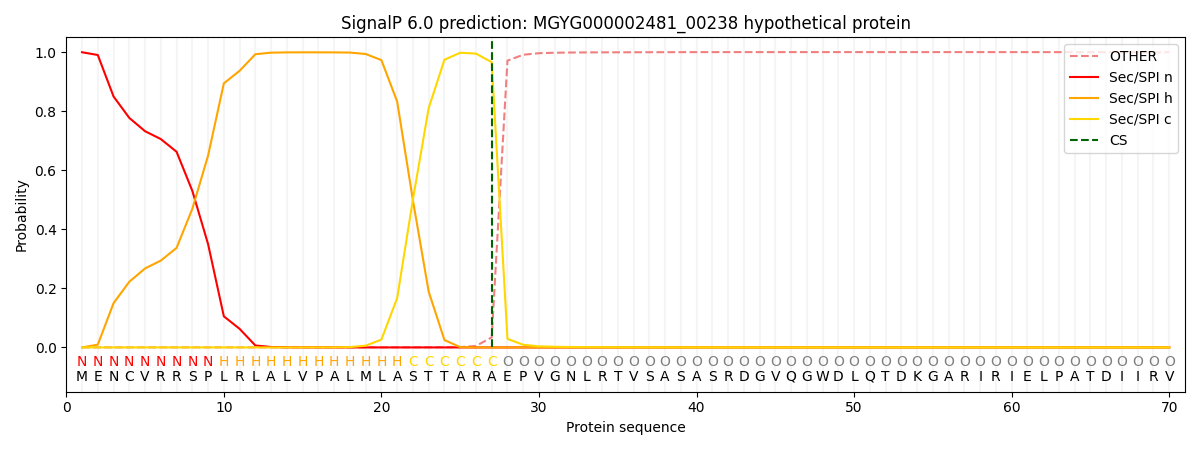
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001426 | 0.996953 | 0.000540 | 0.000610 | 0.000266 | 0.000200 |
