You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000642_00183
You are here: Home > Sequence: MGYG000000642_00183
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
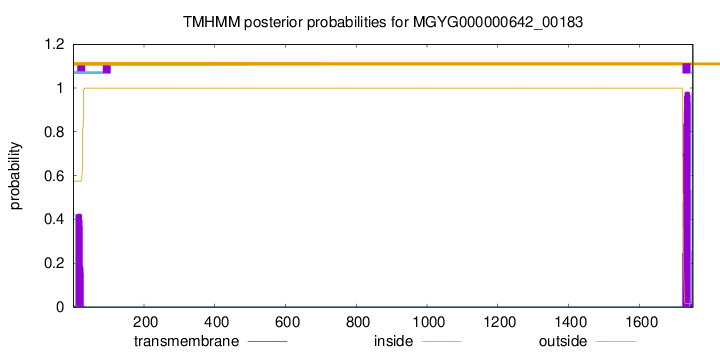
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Arcanobacterium_A; | |||||||||||
| CAZyme ID | MGYG000000642_00183 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 205799; End: 211057 Strand: - | |||||||||||
Full Sequence Download help
| MGKFGKALAS ITGAALGLSA ITMSILEVPQ ANAAEGEFTH TTVVKYGDAS ITNYRIPAVI | 60 |
| KLKNGDLLVS YDARPANGDA PAPNSIMQKR STDGGKTWGE QTTIAAGKGT TMNAPDKEGF | 120 |
| SDPAYIYDET TNTIFNFHVH SYDAGCNQNG STGTDEFTDR NVQGVSLSVS TDNGVTWKER | 180 |
| HGSTEDTDRG LTADVKQPGY PCGFASSGHG IQLKHQTGAN ESKNGRLITQ FIAGKGNEWA | 240 |
| AYSVYSDDHG KTWKRGATIG TDMNENKLVE LSDGTLLLNS RMSSKGGYRH IAKSTDGGET | 300 |
| WSSLELERSL IDSKTPGRNA SIVRMYPEAS ASDPKSKELL YSGMAESGKT LAVSYSYDDG | 360 |
| ATWPIRKLYW NNKAAYSDLV VIDAEKGQYG VAFEEDHQVG NNWFETIHFG TFDKTWLNPV | 420 |
| KVSLADATIE AEAGTTVEIP ITIKNDDSGD TKIPVGTPVK LNLPQGWTME ESVLAQDLYP | 480 |
| NNSETVTLRV SIPANASQQT YYFDSVLESS NIKLRGDVNV NVTNSNIVTD PTKSVGVTFE | 540 |
| LTNPKSDGTW AVGEKMNFTV NVTNMMGQDK SFTATASNLD NGWEGCKWRS LSAGATLPCA | 600 |
| NRTHTVTQAD IDAGGFTPSI TFKYQNTGYS GAATQLEAVQ GKQSPVYARH VKITDLKVKG | 660 |
| TKNSYAVGDK ITFTPTVQNI GNTPIDVQLT GENFTLSCQN ENGKLAKDAT FSCTSSDYSL | 720 |
| KSADIAQGNF TPKVKVKVTD GTKVLQEFTY AGESIPLPSD DPIATKLPQT IPSLDSWKAN | 780 |
| LDANAVFELT AQTKVYYPAG FESQAKIIAQ ELNAYLKAAG LAETVTAQAS DGAKSEINDI | 840 |
| TISIDEAQKT KLGEEGYQLT IGETGVQISA AAKRGAFWGT RTVSQMLRQQ LKLPFGNATD | 900 |
| KPQQTERAVT LCACRIKNQP DYIERMLTDM ADLRMNQVIL TAKMQSNKEP VTNTYSYYSK | 960 |
| EELKDIVAFA DTLGIEIVPQ MGAPGHMDPK IHNLPQYQLK KTDGTLDSDR LDITNPEAVA | 1020 |
| FYKRQIDDYL DVFPSKYWHM AGDEYMYRSS YSNYPVFGEK GKDGARLFVE FVNDINKYVK | 1080 |
| SKGKTLRMWN DGLNASNIDQ LDKDIVIDMW QNGLSPQSLI DKGHKVNNTN YDLYFSREMG | 1140 |
| WRIQNNGAAT IYNDRGFTAG TFKSGKVADN NTNNLGIKLS IWPDTSIYQT ENEVEMETFD | 1200 |
| AFRLVSQIGW SGSRPWKDGA TFVDFAKKLG RNPLWENVNR KPLENGTYAF TAEGLRLDGS | 1260 |
| ENREPDAIAY SDDQWEIKST YDHYYQLKSK ASGKCLTMDV GKHNLNGSKR QYATTDLTVV | 1320 |
| NEVGARPALV DCVADMDVKY HTGTWKNNSS TSIRNNQKWQ IIADNNGRYI VRHALTNMDL | 1380 |
| AISTGEEKHV DLGNVGDMDD NAKSRPMPTD LIQLPSDMTN TRWSITPADR DAAPNMQKPH | 1440 |
| GKGVSVANLP WYDGSTIGWA DDGLIRVNRT LLGSPLLIGG EKYRSGIGTH ASSKITVHLG | 1500 |
| KNCTSFSAKV GIDDTQKDTK NKNADGVIFR VLGADGSELG ASSQQKPGEK AQELKVSVEN | 1560 |
| QEKITLFADA GNSNGNDHAD WADAVVNCNE MDADKYQPAY ADAQAKAGET ATTAVPSFTA | 1620 |
| SEGKAKAKKF EFAETAPANM SIAEDSGAIT WKVPSDHVNE TISVSVKVTY EDGSNEIVTA | 1680 |
| NFQVVAAMPT PQPEQQPGED NPSEKESPQK PTEPIVNKQL QKTGSATAGL IALAVLLSVA | 1740 |
| GGALLTSKKR IW | 1752 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 42 | 406 | 2.7e-74 | 0.9532163742690059 |
| GH20 | 906 | 1212 | 1.3e-51 | 0.9525222551928784 |
| CBM51 | 1446 | 1585 | 8e-32 | 0.9701492537313433 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 5.85e-84 | 41 | 413 | 1 | 339 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| cd06564 | GH20_DspB_LnbB-like | 5.95e-62 | 906 | 1212 | 2 | 326 | Glycosyl hydrolase family 20 (GH20) catalytic domain of dispersin B (DspB), lacto-N-biosidase (LnbB) and related proteins. Dispersin B is a soluble beta-N-acetylglucosamidase found in bacteria that hydrolyzes the beta-1,6-linkages of PGA (poly-beta-(1,6)-N-acetylglucosamine), a major component of the extracellular polysaccharide matrix. Lacto-N-biosidase hydrolyzes lacto-N-biose (LNB) type I oligosaccharides at the nonreducing terminus to produce lacto-N-biose as part of the GNB/LNB (galacto-N-biose/lacto-N-biose I) degradation pathway. The lacto-N-biosidase from Bifidobacterium bifidum has this GH20 domain, a carbohydrate binding module 32, and a bacterial immunoglobulin-like domain 2, as well as a YSIRK signal peptide and a G5 membrane anchor at the N and C termini, respectively. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| pfam08305 | NPCBM | 5.29e-33 | 1444 | 1586 | 3 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| pfam00728 | Glyco_hydro_20 | 8.90e-26 | 920 | 1135 | 17 | 267 | Glycosyl hydrolase family 20, catalytic domain. This domain has a TIM barrel fold. |
| cd06563 | GH20_chitobiase-like | 4.99e-25 | 957 | 1229 | 82 | 357 | The chitobiase of Serratia marcescens is a beta-N-1,4-acetylhexosaminidase with a glycosyl hydrolase family 20 (GH20) domain that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. This GH20 domain family includes an N-acetylglucosamidase (GlcNAcase A) from Pseudoalteromonas piscicida and an N-acetylhexosaminidase (SpHex) from Streptomyces plicatus. SpHex lacks the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWG03116.1 | 6.76e-163 | 724 | 1539 | 264 | 1078 |
| AZR04729.1 | 6.76e-163 | 724 | 1539 | 264 | 1078 |
| AZR07627.1 | 6.76e-163 | 724 | 1539 | 264 | 1078 |
| AWG15845.1 | 6.76e-163 | 724 | 1539 | 264 | 1078 |
| QIU85951.1 | 5.43e-161 | 607 | 1539 | 164 | 1072 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4H04_A | 1.85e-134 | 762 | 1420 | 28 | 643 | Lacto-N-biosidasefrom Bifidobacterium bifidum [Bifidobacterium bifidum JCM 1254],4H04_B Lacto-N-biosidase from Bifidobacterium bifidum [Bifidobacterium bifidum JCM 1254],4JAW_A Crystal Structure of Lacto-N-Biosidase from Bifidobacterium bifidum complexed with LNB-thiazoline [Bifidobacterium bifidum JCM 1254],4JAW_B Crystal Structure of Lacto-N-Biosidase from Bifidobacterium bifidum complexed with LNB-thiazoline [Bifidobacterium bifidum JCM 1254],5BXP_A LNBase in complex with LNB-LOGNAc [Bifidobacterium bifidum JCM 1254],5BXP_B LNBase in complex with LNB-LOGNAc [Bifidobacterium bifidum JCM 1254],5BXR_A LNBase in complex with LNB-NHAcDNJ [Bifidobacterium bifidum JCM 1254],5BXR_B LNBase in complex with LNB-NHAcDNJ [Bifidobacterium bifidum JCM 1254],5BXS_A LNBase in complex with LNB-NHAcCAS [Bifidobacterium bifidum JCM 1254],5BXS_B LNBase in complex with LNB-NHAcCAS [Bifidobacterium bifidum JCM 1254],5BXT_A LNBase in complex with LNB-NHAcAUS [Bifidobacterium bifidum JCM 1254],5BXT_B LNBase in complex with LNB-NHAcAUS [Bifidobacterium bifidum JCM 1254] |
| 1EUT_A | 2.60e-86 | 38 | 510 | 9 | 447 | Sialidase,Large 68kd Form, Complexed With Galactose [Micromonospora viridifaciens],1EUU_A Sialidase Or Neuraminidase, Large 68kd Form [Micromonospora viridifaciens] |
| 1WCQ_A | 7.93e-86 | 38 | 510 | 5 | 443 | Mutagenesisof the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_B Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_C Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens] |
| 2BZD_A | 1.46e-85 | 38 | 510 | 5 | 443 | Galactoserecognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_B Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_C Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens] |
| 1W8N_A | 1.98e-85 | 38 | 510 | 5 | 443 | Contributionof the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. [Micromonospora viridifaciens],1W8O_A Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02834 | 1.63e-85 | 24 | 510 | 37 | 489 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| B2UPR7 | 1.19e-20 | 832 | 1130 | 164 | 485 | Beta-hexosaminidase Amuc_2136 OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=Amuc_2136 PE=1 SV=1 |
| P49610 | 4.28e-16 | 921 | 1135 | 226 | 470 | Beta-N-acetylhexosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=strH PE=1 SV=2 |
| A9WNA0 | 5.82e-14 | 1436 | 1588 | 1155 | 1305 | Putative endo-alpha-N-acetylgalactosaminidase OS=Renibacterium salmoninarum (strain ATCC 33209 / DSM 20767 / JCM 11484 / NBRC 15589 / NCIMB 2235) OX=288705 GN=RSal33209_1326 PE=3 SV=2 |
| P13723 | 1.05e-13 | 834 | 1126 | 80 | 391 | Beta-hexosaminidase subunit A1 OS=Dictyostelium discoideum OX=44689 GN=hexa1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000424 | 0.998725 | 0.000191 | 0.000250 | 0.000220 | 0.000180 |