You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003059_00167
You are here: Home > Sequence: MGYG000003059_00167
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
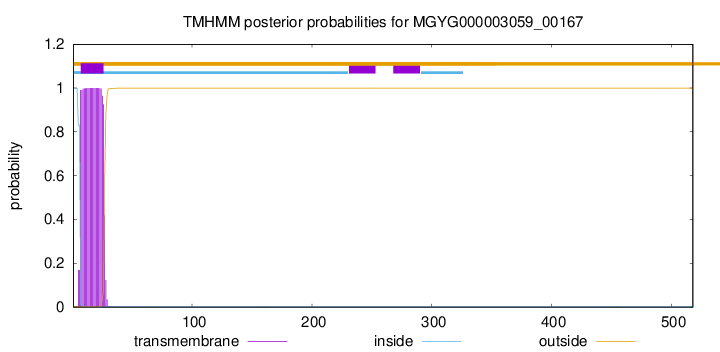
TMHMM annotations
Basic Information help
| Species | CAG-180 sp900545625 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; CAG-180; CAG-180 sp900545625 | |||||||||||
| CAZyme ID | MGYG000003059_00167 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | Sialidase B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 21077; End: 22633 Strand: - | |||||||||||
Full Sequence Download help
| MRKVIKLLFS CLGAAAVFVL VLGFFITPKA GGAPNIAVDG VKPFSSASVI NCNYYRIPAL | 60 |
| LTTTDGTVLA AIDARFGGTH DSPNNIDTAV SISTDSGKSW SEPQLVFQFD DFENTDDFLK | 120 |
| ENGTYTTQNS ASVIDPALLQ DTQTGRIFML VDAFPHGAGA FAAQVGSGYT EINGEQALLL | 180 |
| RKQGEEDFAY TLRADGAIYD ANGTKTAYTV NENGELSENG VPLTVQQKTQ KYWYNLPFNL | 240 |
| NTRKEIPMHI FYRDSQFQPL STSYLYLLYS DDNGETWSAP IDLNAQVKPD DVGFFGVCPG | 300 |
| RGLQIQNGTY AGRLLFCAYT LDPESGEQRF MTIFSDDGGS TWQAGEFVAL TDEIRSMSET | 360 |
| QLIELPDGSL QAYSRTTNGF VSVAFSTDGG ATWQDPQLVE NLSLTSGSGC QISVINCSQK | 420 |
| IDGKDVVLLS APAGDSRRNG YIYVGLVEQN GDAAGYVVNW TYKKEITNAD TYFAYSCLTE | 480 |
| LPDGNIGLLY EQANTPQNVD TVVFASFSLS ELCEEEIQ | 518 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 51 | 507 | 7.2e-92 | 0.9473684210526315 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 1.49e-88 | 53 | 508 | 12 | 339 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| COG4409 | NanH | 3.91e-62 | 53 | 495 | 273 | 704 | Neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| pfam13088 | BNR_2 | 2.05e-19 | 261 | 398 | 78 | 203 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANU76745.2 | 8.28e-103 | 44 | 492 | 233 | 682 |
| QJU17595.1 | 8.28e-103 | 44 | 492 | 233 | 682 |
| ASU31741.1 | 8.28e-103 | 44 | 492 | 233 | 682 |
| QQQ95451.1 | 8.28e-103 | 44 | 492 | 233 | 682 |
| ETD19277.1 | 2.61e-100 | 44 | 492 | 245 | 694 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4X6K_A | 8.51e-105 | 40 | 492 | 4 | 457 | Crystalstructure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with Siastatin B [[Ruminococcus] gnavus CC55_001C] |
| 4X47_A | 9.30e-105 | 40 | 492 | 7 | 460 | Crystalstructure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with Neu5Ac2en [[Ruminococcus] gnavus ATCC 29149],4X49_A Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with oseltamivir carboxylate [[Ruminococcus] gnavus ATCC 29149],4X4A_A Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with 2,7-Anhydro-Neu5Ac [[Ruminococcus] gnavus ATCC 29149] |
| 1SLI_A | 1.66e-102 | 33 | 512 | 190 | 675 | LeechIntramolecular Trans-Sialidase Complexed With Dana [Macrobdella decora],1SLL_A Sialidase L From Leech Macrobdella Decora [Macrobdella decora],2SLI_A Leech Intramolecular Trans-Sialidase Complexed With 2,7- Anhydro-Neu5ac, The Reaction Product [Macrobdella decora],3SLI_A Leech Intramolecular Trans-Sialidase Complexed With 2,7- Anhydro-Neu5ac Prepared By Soaking With 3'-Sialyllactose [Macrobdella decora],4SLI_A Leech Intramolecular Trans-Sialidase Complexed With 2- Propenyl-Neu5ac, An Inactive Substrate Analogue [Macrobdella decora] |
| 5TSP_A | 6.65e-97 | 50 | 491 | 19 | 430 | Crystalstructure of the catalytic domain of Clostridium perfringens neuraminidase (NanI) in complex with a CHES [Clostridium perfringens ATCC 13124],5TSP_B Crystal structure of the catalytic domain of Clostridium perfringens neuraminidase (NanI) in complex with a CHES [Clostridium perfringens ATCC 13124] |
| 2BF6_A | 3.66e-96 | 50 | 491 | 18 | 429 | AtomicResolution Structure of the bacterial sialidase NanI from Clostridium perfringens in complex with alpha-Sialic Acid (Neu5Ac). [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q27701 | 6.54e-101 | 33 | 512 | 270 | 755 | Anhydrosialidase OS=Macrobdella decora OX=6405 PE=1 SV=1 |
| P29767 | 1.80e-93 | 50 | 497 | 390 | 815 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
| Q54727 | 3.08e-76 | 44 | 492 | 233 | 670 | Sialidase B OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=nanB PE=1 SV=2 |
| P62576 | 1.65e-59 | 32 | 517 | 336 | 794 | Sialidase A OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=nanA PE=1 SV=1 |
| P62575 | 1.65e-59 | 32 | 517 | 336 | 794 | Sialidase A OS=Streptococcus pneumoniae OX=1313 GN=nanA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
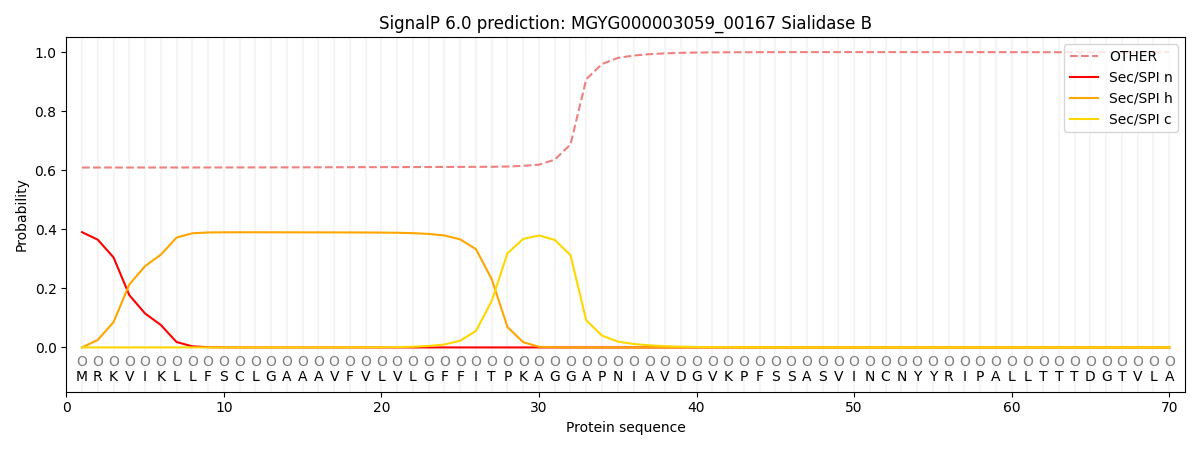
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.618849 | 0.378935 | 0.001279 | 0.000385 | 0.000232 | 0.000320 |