You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004571_00362
You are here: Home > Sequence: MGYG000004571_00362
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA6382 sp900557555 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; UBA6382; UBA6382 sp900557555 | |||||||||||
| CAZyme ID | MGYG000004571_00362 | |||||||||||
| CAZy Family | GH123 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25393; End: 30108 Strand: - | |||||||||||
Full Sequence Download help
| MKKSLSLIIC LLFVTVVLHA QNYTVAHGSD EHITNDIHYP KAVGVKGTQT AEFKLENIAT | 60 |
| APGCAAYFDR TSTVFEVKAG DIITPSVTIN GAWMHSFIFV DWSQNGQFEV NLLGDGPYTE | 120 |
| GDGNELVSYS HYNMASSTVN NGWNSAGEAV GGNVLTPCTF RVPAHLAVGS SYRMRYSVTW | 180 |
| NCARPDGDYP NFISDGGSII DVTLKIVGVA ENVQKYPIDN YEEPSVGDVP PTEQWNALSA | 240 |
| GLHCSWANRN EHYKKHLVPE LTETTVATVT AWKGERANIE AVLFSKEDQG TLRVRMTELR | 300 |
| KNGEGTGIEC ATARFMNYII TDDGRGCGDH NFSLPTWLVP DVIEQPNKAK VVNACETRPV | 360 |
| WCSIEVPRNI DAGTYSTAME VVNEAGDIVK TLDLTIEVVD RALPKAADRK FHLDLWQQPY | 420 |
| AVSRYYEVPR WSDEHIAALR PYLEALGKAG QSVVTTILFY EPWGEQSHDK FSPMIQTTRK | 480 |
| KDGTWAFNYD IFDKYVELCN ECGINRQINC YSMVTWDMKF RYHDEATDTD VDWALAVGTK | 540 |
| DYNEMWDAYL KAFRSHLEKK GWFDKTCIAM DERSESQMLS AYNIIRANGF KMALAGNYHS | 600 |
| SLNDKLYDYC VALGQGGRFT DAERKYRKDN NLLTTVYTCC TESEPNIFSN SLPAEAAFLP | 660 |
| IHVAANGLDG YLHWSWINWD EHPLTDTRFR KFGAGDTYCY YPGNRSSVRF ERLVEGIQQF | 720 |
| EKIQIMKREM AGDATWMETF DKLLGNCKDF FTSGAECAGK VDRLEAFLNG KEVDMPEDVT | 780 |
| GYWKIKVDDT HYAKSAANQT EWETELSAQA GFDLFLIEGD KEACTIKLMG RTNNIGPDNS | 840 |
| GKLFVDQTAT TKYSIVDAAD GKVKLKNNGY WVYVDNGTFT WNATKSTELQ LEFVMPYKKL | 900 |
| ARTILFSTLN GGMNIPPYRI PGITCGKNGR LIASAARLVC GTDPGFGQVD CVVKISDDNG | 960 |
| ITWSEQEIEV AVGDATLINN VKTPMEAAYG DPAVVFDREH NEALVMAVAG CTVYTSPTTN | 1020 |
| RQNPNMIAAI RSLDGGNTWE TPVDQTEHIY GLFDSGKPLA AAFVGGGKVF QSRVVKVGAY | 1080 |
| YRLYAALDAR PGGNRIIYSD DFGRSWKALG GASATPVPDG DEPKCEELPD GRVIVTSRTG | 1140 |
| GGRLMNIYTY SNTRTGAGKW EAQVKATMSG LSASPSSNPT NGEMLIVPAQ RTSDDMMLYV | 1200 |
| ALQSVPTGSG RNNVGIFYKE LADADDIRNV TAFTDAWDGF YQVSSTASAY SSLDLQADDK | 1260 |
| IGFFYEETLT KWGVKPNPVS TSFPTGSGTH NFDGFENVYV ALDLEMITSG KYTVNRNVNR | 1320 |
| GDYLKNYFNS IIDEADMPDD EKDAARAEVA KLSNDPAMEQ IDAIYGLIEG AEPADKWDGK | 1380 |
| VLAFTNVQQN NTERTLYIEN GKLKFASATV ESLGESAQFK CTKRENGKYS LLNENSGLYM | 1440 |
| IWRSSDAQNY GYNNNCGTLT GYNATYCDWV FADASGTKAN TYYIVSKRAD GSTDGSLVIM | 1500 |
| KSGVFDSYGA SVGWNANYSN LFRINIVDLT TGLKVETTAE PSAMDGKYLH NNRVVVIRNG | 1560 |
| VHYDTSGRRH N | 1571 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH123 | 240 | 750 | 2.1e-173 | 0.9572490706319703 |
| GH33 | 909 | 1268 | 5.4e-41 | 0.9239766081871345 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 4.75e-48 | 903 | 1267 | 1 | 324 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| pfam13320 | DUF4091 | 6.07e-19 | 667 | 727 | 1 | 66 | Domain of unknown function (DUF4091). This presumed domain is functionally uncharacterized. This domain family is found in bacteria, archaea and eukaryotes, and is approximately 70 amino acids in length. There is a single completely conserved residue G that may be functionally important. |
| COG4409 | NanH | 1.70e-04 | 901 | 1039 | 258 | 411 | Neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFQ13451.1 | 1.65e-169 | 235 | 789 | 44 | 600 |
| QQR08655.1 | 1.07e-140 | 236 | 769 | 16 | 537 |
| ANU63982.1 | 1.07e-140 | 236 | 769 | 16 | 537 |
| ASB37921.1 | 1.07e-140 | 236 | 769 | 16 | 537 |
| ASM64853.1 | 4.58e-138 | 192 | 744 | 12 | 571 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5L7V_A | 9.57e-115 | 245 | 744 | 29 | 536 | ChainA, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7V_B Chain B, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
| 5L7R_A | 1.50e-114 | 245 | 744 | 44 | 551 | ChainA, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7R_B Chain B, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7U_A Chain A, Glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7U_B Chain B, Glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
| 5FQE_A | 6.03e-60 | 242 | 726 | 30 | 542 | Thedetails of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQE_B The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQF_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQF_B The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FR0_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 5FQG_A | 6.62e-59 | 242 | 726 | 30 | 542 | Thedetails of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQH_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 1EUR_A | 6.37e-13 | 909 | 1138 | 16 | 234 | Sialidase[Micromonospora viridifaciens],1EUS_A Sialidase Complexed With 2-Deoxy-2,3-Dehydro-N- Acetylneuraminic Acid [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02834 | 9.70e-12 | 909 | 1138 | 58 | 276 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
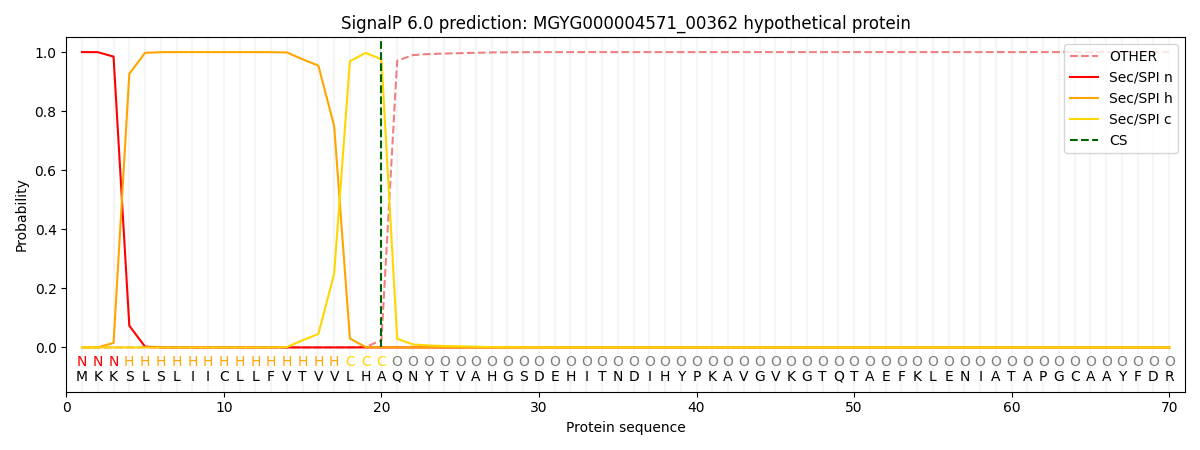
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000248 | 0.999133 | 0.000172 | 0.000152 | 0.000145 | 0.000143 |
