You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002470_00046
You are here: Home > Sequence: MGYG000002470_00046
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides intestinalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides intestinalis | |||||||||||
| CAZyme ID | MGYG000002470_00046 | |||||||||||
| CAZy Family | GH36 | |||||||||||
| CAZyme Description | Alpha-galactosidase AgaA | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 55936; End: 58125 Strand: + | |||||||||||
Full Sequence Download help
| MKKLLVTLFA CATLLYTHAA ENKAIRISTD NTDLVLQVGD NGRLYQTYLG ERLLHEQDLN | 60 |
| NFSWSIHAGA DGSVSRRGWE VYSGSGNEDY FEPAIAITHN DGNPSTILYY VSSSSKAVEG | 120 |
| GMETVINLRD DKYPVDVTLH YVAYPKENVI KTWSEIIHQE KKPVMLSTYA STMLYFNNSA | 180 |
| YYLTEFSSDW AKEAQMSSQQ LQFGKKVIDT KLGSRAAMHT HPFFEVGLDQ PVREDQGSVL | 240 |
| MGTLGWIGNF RFIFEVDNVG NLRVIPGINP YASNYELKPN EVFTTPEFIF TLSNDGAGKG | 300 |
| SRNLHEWARN YSLKDGKGSR LTLLNNWENT GFNFNQEILT ELMKEAKHLG VDMFLLDDGW | 360 |
| FANKYPRKND HAGLGDWDVN HNKLPGGIPA LVKAAKDADV KFGIWIEPEM VNPKSELFEK | 420 |
| HPDWAIMLPN RDTYYYRNQL VLDLSNPKVQ DYVYSVVENI MKENPEVAFF KWDCNSPITN | 480 |
| IYSPYLKNKQ GQLYIDHVRG IYNVLKRVKE NYPDVPMMLC SGGGARCDYE ALKYFTEFWC | 540 |
| SDNTDPVERI YIQWGFSQFF PAKAMDAHVT SWNKATSVKF RTDVASMCKL GFDIGLKELT | 600 |
| ADELAYCQTA VANWKRLQNA IMDGDQYRLV SPYEGNHMAL NYVSKDTNKA VLFAYDIHPR | 660 |
| FQEKLMAVKL QGLDPNKQYK VEEINLMPSV TSKLGSNGKV FSGDYLMKVG LNVFGLATTQ | 720 |
| SHVIELTAQ | 729 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH36 | 25 | 712 | 1.3e-207 | 0.9956395348837209 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02065 | Melibiase | 4.33e-111 | 283 | 624 | 2 | 347 | Melibiase. Glycoside hydrolase families GH27, GH31 and GH36 form the glycoside hydrolase clan GH-D. Glycoside hydrolase family 36 can be split into 11 families, GH36A to GH36K. This family includes enzymes from GH36A-B and GH36D-K and from GH27. |
| COG3345 | GalA | 3.78e-108 | 22 | 713 | 1 | 667 | Alpha-galactosidase [Carbohydrate transport and metabolism]. |
| cd14791 | GH36 | 2.48e-104 | 320 | 616 | 2 | 299 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| pfam16875 | Glyco_hydro_36N | 4.81e-71 | 42 | 278 | 1 | 256 | Glycosyl hydrolase family 36 N-terminal domain. This domain is found at the N-terminus of many family 36 glycoside hydrolases. It has a beta-supersandwich fold. |
| pfam16874 | Glyco_hydro_36C | 1.95e-22 | 637 | 726 | 1 | 78 | Glycosyl hydrolase family 36 C-terminal domain. This domain is found at the C-terminus of many family 36 glycoside hydrolases. It has a beta-sandwich structure with a Greek key motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO71274.1 | 0.0 | 1 | 729 | 1 | 729 |
| ALJ58592.1 | 0.0 | 1 | 729 | 1 | 729 |
| QUT90297.1 | 0.0 | 1 | 729 | 1 | 729 |
| QRO24673.1 | 0.0 | 1 | 728 | 1 | 728 |
| QCD36631.1 | 0.0 | 18 | 728 | 20 | 730 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4FNQ_A | 6.37e-109 | 17 | 711 | 4 | 708 | Crystalstructure of GH36 alpha-galactosidase AgaB from Geobacillus stearothermophilus [Geobacillus stearothermophilus] |
| 4FNR_A | 2.61e-107 | 80 | 711 | 74 | 708 | Crystalstructure of GH36 alpha-galactosidase AgaA from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNR_B Crystal structure of GH36 alpha-galactosidase AgaA from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNR_C Crystal structure of GH36 alpha-galactosidase AgaA from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNR_D Crystal structure of GH36 alpha-galactosidase AgaA from Geobacillus stearothermophilus [Geobacillus stearothermophilus] |
| 4FNP_A | 2.08e-105 | 80 | 711 | 74 | 708 | Crystalstructure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNP_B Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNP_C Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNP_D Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNS_A Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin [Geobacillus stearothermophilus],4FNS_B Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin [Geobacillus stearothermophilus],4FNS_C Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin [Geobacillus stearothermophilus],4FNS_D Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin [Geobacillus stearothermophilus] |
| 4FNU_A | 2.08e-105 | 80 | 711 | 74 | 708 | Crystalstructure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose [Geobacillus stearothermophilus],4FNU_B Crystal structure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose [Geobacillus stearothermophilus],4FNU_C Crystal structure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose [Geobacillus stearothermophilus],4FNU_D Crystal structure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose [Geobacillus stearothermophilus] |
| 4FNT_A | 2.92e-105 | 80 | 711 | 74 | 708 | Crystalstructure of GH36 alpha-galactosidase AgaA A355E D548N from Geobacillus stearothermophilus in complex with raffinose [Geobacillus stearothermophilus],4FNT_B Crystal structure of GH36 alpha-galactosidase AgaA A355E D548N from Geobacillus stearothermophilus in complex with raffinose [Geobacillus stearothermophilus],4FNT_C Crystal structure of GH36 alpha-galactosidase AgaA A355E D548N from Geobacillus stearothermophilus in complex with raffinose [Geobacillus stearothermophilus],4FNT_D Crystal structure of GH36 alpha-galactosidase AgaA A355E D548N from Geobacillus stearothermophilus in complex with raffinose [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9ALJ4 | 1.43e-106 | 80 | 711 | 74 | 708 | Alpha-galactosidase AgaA OS=Geobacillus stearothermophilus OX=1422 GN=agaA PE=1 SV=1 |
| Q0CVH2 | 1.26e-104 | 24 | 711 | 39 | 729 | Probable alpha-galactosidase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=aglC PE=3 SV=1 |
| Q92457 | 4.73e-104 | 86 | 729 | 97 | 745 | Alpha-galactosidase 2 OS=Hypocrea jecorina OX=51453 GN=agl2 PE=1 SV=1 |
| Q2TW69 | 7.84e-103 | 86 | 729 | 103 | 751 | Probable alpha-galactosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=aglC PE=3 SV=1 |
| B8NWY6 | 7.84e-103 | 86 | 729 | 103 | 751 | Probable alpha-galactosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=aglC PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
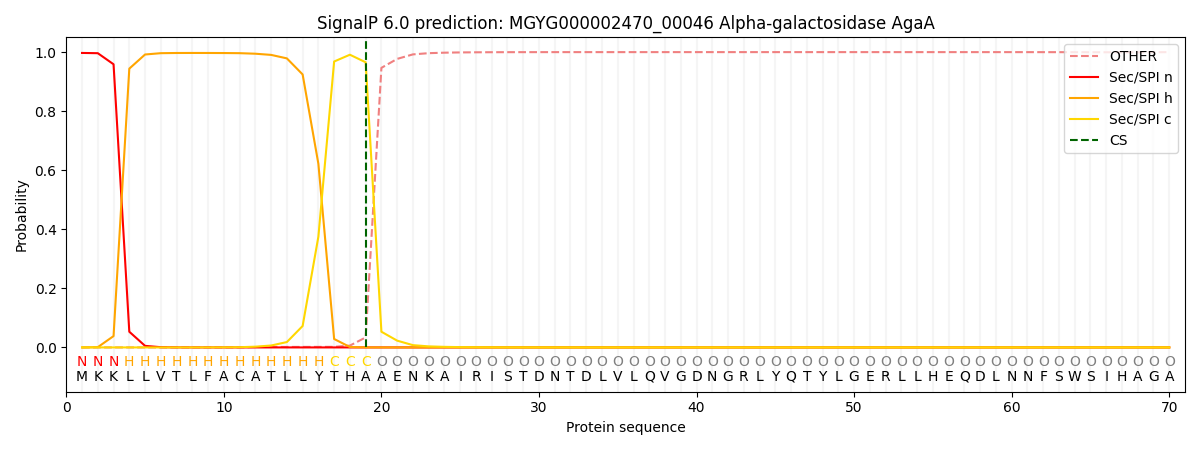
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000535 | 0.993926 | 0.004697 | 0.000289 | 0.000279 | 0.000251 |
