You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002762_00107
You are here: Home > Sequence: MGYG000002762_00107
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-485 sp900554845 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900554845 | |||||||||||
| CAZyme ID | MGYG000002762_00107 | |||||||||||
| CAZy Family | GH36 | |||||||||||
| CAZyme Description | Alpha-galactosidase AgaA | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18294; End: 20489 Strand: - | |||||||||||
Full Sequence Download help
| MNHRTLSAAL LLLAAAPALD AEIINIVTDN NSLVLDAVKG GPLRYVYYGT RLSDAEANSV | 60 |
| WDANIPACEA YPFYGSYPEH ESAMAMTHAD GNMTMDMAVE SVSKTTVDGQ PVTVVTMKDR | 120 |
| YYPLTVKLNY RTYPKDDIIE MWTETTNGEK KPVVLRQFAS GSLPVRCEDV WIQNYHGGWA | 180 |
| NEARVNDEPL RPGMKVIKNR DGARNAHTSR GEIMLSLDGK AREQTGRVIG AAVEYGGNYK | 240 |
| LRIDTDESLH HRLFAGMDEE NSWYNLKPGE TFVTPPLALT FSSDGMGTVS RRFHRWGREN | 300 |
| RLANGDKARK ILLNSWEGVY FDINEEGMDR MMKDIASMGG ELFVMDDGWF GAKYPRKNDS | 360 |
| SSLGDWIVDT NKLPNGINGL LESAKKNGVK FGIWIEPEMT NTISEFYEKH PELVVKAKNR | 420 |
| DVVKGRGGTQ LVLDMGNPKA QEVVFQVVDT LLTKYPDIDY IKWDANAPIM SHGSQYQTAD | 480 |
| NQSHLYIDYH KGLVKTLERI RAKYPDVTIQ LCASGGGRVN WGLLPYFDEF WTSDNTDALQ | 540 |
| RIYMQWGTSS FFPAIAMGSH ISATPNHTTF RSVPIKFRAD VAMSGRLGME IQPANMNEED | 600 |
| KALCKQAIEA YKGIRDIVQF GDLYRLLSPY DEKGAASLMY VTPEKDDAVF FWWKTNPNYG | 660 |
| EKIPRVRMAG LQPERLYTVT ELNRIDNKPL PFEGKQFTGK FLMESGLEIP VSHDVDWNKK | 720 |
| MDYSSRVLRI K | 731 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH36 | 24 | 708 | 1.9e-203 | 0.9956395348837209 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02065 | Melibiase | 1.19e-120 | 271 | 621 | 1 | 347 | Melibiase. Glycoside hydrolase families GH27, GH31 and GH36 form the glycoside hydrolase clan GH-D. Glycoside hydrolase family 36 can be split into 11 families, GH36A to GH36K. This family includes enzymes from GH36A-B and GH36D-K and from GH27. |
| cd14791 | GH36 | 2.51e-108 | 308 | 612 | 1 | 298 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| COG3345 | GalA | 8.24e-106 | 41 | 686 | 19 | 655 | Alpha-galactosidase [Carbohydrate transport and metabolism]. |
| pfam16875 | Glyco_hydro_36N | 2.08e-37 | 41 | 267 | 1 | 256 | Glycosyl hydrolase family 36 N-terminal domain. This domain is found at the N-terminus of many family 36 glycoside hydrolases. It has a beta-supersandwich fold. |
| pfam16874 | Glyco_hydro_36C | 1.20e-18 | 636 | 730 | 2 | 78 | Glycosyl hydrolase family 36 C-terminal domain. This domain is found at the C-terminus of many family 36 glycoside hydrolases. It has a beta-sandwich structure with a Greek key motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCP71829.1 | 0.0 | 8 | 730 | 10 | 733 |
| QCD38146.1 | 0.0 | 8 | 730 | 10 | 733 |
| ADY36266.1 | 0.0 | 22 | 731 | 26 | 735 |
| QCD39023.1 | 0.0 | 1 | 728 | 1 | 732 |
| QCP72715.1 | 0.0 | 1 | 728 | 1 | 732 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4FNQ_A | 5.18e-118 | 32 | 694 | 20 | 707 | Crystalstructure of GH36 alpha-galactosidase AgaB from Geobacillus stearothermophilus [Geobacillus stearothermophilus] |
| 4FNR_A | 3.03e-116 | 32 | 731 | 20 | 725 | Crystalstructure of GH36 alpha-galactosidase AgaA from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNR_B Crystal structure of GH36 alpha-galactosidase AgaA from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNR_C Crystal structure of GH36 alpha-galactosidase AgaA from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNR_D Crystal structure of GH36 alpha-galactosidase AgaA from Geobacillus stearothermophilus [Geobacillus stearothermophilus] |
| 4FNP_A | 3.25e-115 | 32 | 694 | 20 | 707 | Crystalstructure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNP_B Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNP_C Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNP_D Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNS_A Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin [Geobacillus stearothermophilus],4FNS_B Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin [Geobacillus stearothermophilus],4FNS_C Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin [Geobacillus stearothermophilus],4FNS_D Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin [Geobacillus stearothermophilus] |
| 4FNT_A | 4.56e-115 | 32 | 694 | 20 | 707 | Crystalstructure of GH36 alpha-galactosidase AgaA A355E D548N from Geobacillus stearothermophilus in complex with raffinose [Geobacillus stearothermophilus],4FNT_B Crystal structure of GH36 alpha-galactosidase AgaA A355E D548N from Geobacillus stearothermophilus in complex with raffinose [Geobacillus stearothermophilus],4FNT_C Crystal structure of GH36 alpha-galactosidase AgaA A355E D548N from Geobacillus stearothermophilus in complex with raffinose [Geobacillus stearothermophilus],4FNT_D Crystal structure of GH36 alpha-galactosidase AgaA A355E D548N from Geobacillus stearothermophilus in complex with raffinose [Geobacillus stearothermophilus] |
| 4FNU_A | 8.98e-115 | 32 | 731 | 20 | 725 | Crystalstructure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose [Geobacillus stearothermophilus],4FNU_B Crystal structure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose [Geobacillus stearothermophilus],4FNU_C Crystal structure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose [Geobacillus stearothermophilus],4FNU_D Crystal structure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9ALJ4 | 1.66e-115 | 32 | 731 | 20 | 725 | Alpha-galactosidase AgaA OS=Geobacillus stearothermophilus OX=1422 GN=agaA PE=1 SV=1 |
| Q5AU92 | 4.27e-114 | 114 | 726 | 156 | 749 | Alpha-galactosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=aglC PE=1 SV=1 |
| Q9UUZ4 | 3.22e-112 | 98 | 726 | 135 | 746 | Alpha-galactosidase C OS=Aspergillus niger OX=5061 GN=aglC PE=1 SV=1 |
| Q0CEF5 | 2.94e-110 | 114 | 728 | 130 | 720 | Probable alpha-galactosidase G OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=aglG PE=3 SV=1 |
| Q0CVH2 | 7.15e-110 | 114 | 715 | 152 | 737 | Probable alpha-galactosidase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=aglC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
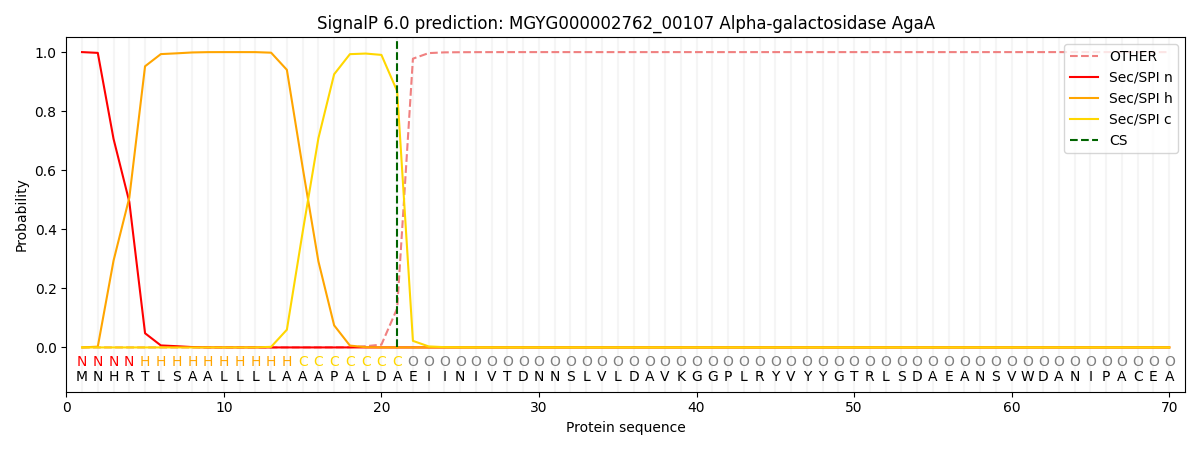
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000267 | 0.999086 | 0.000145 | 0.000180 | 0.000154 | 0.000147 |
