You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002497_00183
You are here: Home > Sequence: MGYG000002497_00183
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
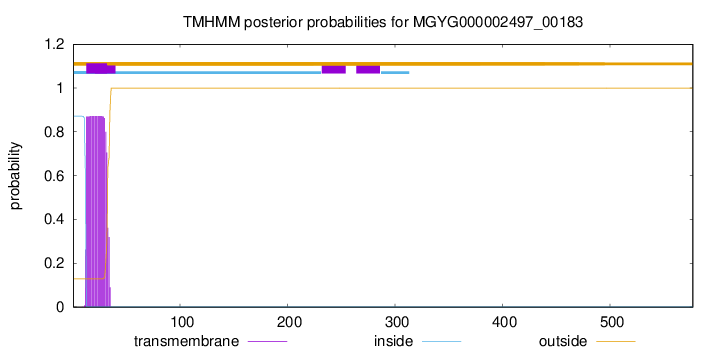
TMHMM annotations
Basic Information help
| Species | Klebsiella aerogenes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Klebsiella; Klebsiella aerogenes | |||||||||||
| CAZyme ID | MGYG000002497_00183 | |||||||||||
| CAZy Family | GH37 | |||||||||||
| CAZyme Description | Periplasmic trehalase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 171736; End: 173469 Strand: - | |||||||||||
Full Sequence Download help
| MTRSVLRRPE TLTYLIRLAL GGAILSLATF AAQADDNTSP VPQSPDELLG PLFNDVQSAK | 60 |
| LFPDQKTFAD AVPNSDPLMI LADYRMQKSQ ASFDLRHFVE INFTLPRDND QYVPPKGQTL | 120 |
| RQHIDGLWPV LTRSTVEVEK WDSLLPLPKP YVVPGGRFRE VYYWDSYFTM LGLAESGHWD | 180 |
| KVEDMVANFA AEIDRWGHIP NGNRSYYLSR SQPPFFSFMV ELLASHDGDR ALKTWLPQME | 240 |
| KEYRYWMEGA DALTPGKANK RVVRMEDGAL LNRYWDDNDT PRPESWLDDV TTAKNNPNRP | 300 |
| ATEIYRDLRS AAASGWDFSS RWMDNPQQLG TIRTTSILPV DLNALMFHME KTIARASKAA | 360 |
| GDSAKAGQYD ALANARQKAL EKYLWNDKEG WYADYDLKSH KVRNQLTAAA LFPLYVKAAS | 420 |
| RERAAKVAAA AESRLLKPGG LTTTTVNSGQ QWDAPNGWAP LQWVAVEGLQ NYGQKKVAME | 480 |
| VTWRFLSNVQ HTYDSKQKLV EKYDVSSTGT GGGGGEYPLQ DGFGWTNGVT LKMLDLVCPQ | 540 |
| EKPCDALPAT RPTQTESATG QQPAANDAVA AAEKKAQ | 577 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH37 | 61 | 536 | 3.7e-189 | 0.9959266802443992 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK13270 | treF | 0.0 | 52 | 536 | 62 | 545 | alpha,alpha-trehalase TreF. |
| PRK13272 | treA | 0.0 | 8 | 540 | 4 | 539 | alpha,alpha-trehalase TreA. |
| COG1626 | TreA | 0.0 | 38 | 543 | 47 | 558 | Neutral trehalase [Carbohydrate transport and metabolism]. |
| PRK13271 | treA | 0.0 | 1 | 563 | 1 | 562 | alpha,alpha-trehalase TreA. |
| pfam01204 | Trehalase | 0.0 | 60 | 536 | 1 | 507 | Trehalase. Trehalase (EC:3.2.1.28) is known to recycle trehalose to glucose. Trehalose is a physiological hallmark of heat-shock response in yeast and protects of proteins and membranes against a variety of stresses. This family is found in conjunction with pfam07492 in fungi. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AKK79885.1 | 0.0 | 1 | 577 | 1 | 577 |
| QRZ88782.1 | 0.0 | 1 | 577 | 1 | 577 |
| QRZ85718.1 | 0.0 | 1 | 577 | 1 | 577 |
| QFI16813.1 | 0.0 | 1 | 577 | 1 | 577 |
| AWD04164.1 | 0.0 | 1 | 577 | 1 | 577 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JG0_A | 0.0 | 36 | 568 | 1 | 531 | Family37 trehalase from Escherichia coli in complex with 1- thiatrehazolin [Escherichia coli K-12],2JJB_A Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2JJB_B Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2JJB_C Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2JJB_D Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2WYN_A Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12],2WYN_B Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12],2WYN_C Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12],2WYN_D Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12] |
| 5Z66_A | 0.0 | 32 | 570 | 34 | 561 | Structureof periplasmic trehalase from Diamondback moth gut bacteria complexed with validoxylamine [Enterobacter cloacae],5Z6H_A Structure of periplasmic trehalase from Diamondback moth gut bacteria in the apo form [Enterobacter cloacae],5Z6H_B Structure of periplasmic trehalase from Diamondback moth gut bacteria in the apo form [Enterobacter cloacae] |
| 2JF4_A | 3.94e-317 | 36 | 568 | 1 | 531 | Family37 trehalase from Escherichia coli in complex with validoxylamine [Escherichia coli K-12] |
| 7E9U_A | 3.14e-79 | 122 | 535 | 123 | 552 | ChainA, Trehalase [Arabidopsis thaliana],7E9U_B Chain B, Trehalase [Arabidopsis thaliana] |
| 7E9X_A | 4.63e-78 | 122 | 535 | 123 | 552 | ChainA, Trehalase [Arabidopsis thaliana],7E9X_B Chain B, Trehalase [Arabidopsis thaliana],7E9X_C Chain C, Trehalase [Arabidopsis thaliana],7E9X_D Chain D, Trehalase [Arabidopsis thaliana],7EAW_A Chain A, Trehalase [Arabidopsis thaliana],7EAW_B Chain B, Trehalase [Arabidopsis thaliana] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| C0Q337 | 0.0 | 6 | 565 | 6 | 567 | Periplasmic trehalase OS=Salmonella paratyphi C (strain RKS4594) OX=476213 GN=treA PE=3 SV=1 |
| P59765 | 0.0 | 6 | 565 | 6 | 567 | Putative periplasmic trehalase OS=Salmonella typhi OX=90370 GN=treA PE=5 SV=2 |
| P13482 | 0.0 | 25 | 568 | 21 | 561 | Periplasmic trehalase OS=Escherichia coli (strain K12) OX=83333 GN=treA PE=1 SV=1 |
| C4ZTN8 | 0.0 | 25 | 568 | 21 | 561 | Periplasmic trehalase OS=Escherichia coli (strain K12 / MC4100 / BW2952) OX=595496 GN=treA PE=3 SV=1 |
| Q0TIH3 | 0.0 | 25 | 559 | 21 | 553 | Periplasmic trehalase OS=Escherichia coli O6:K15:H31 (strain 536 / UPEC) OX=362663 GN=treA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
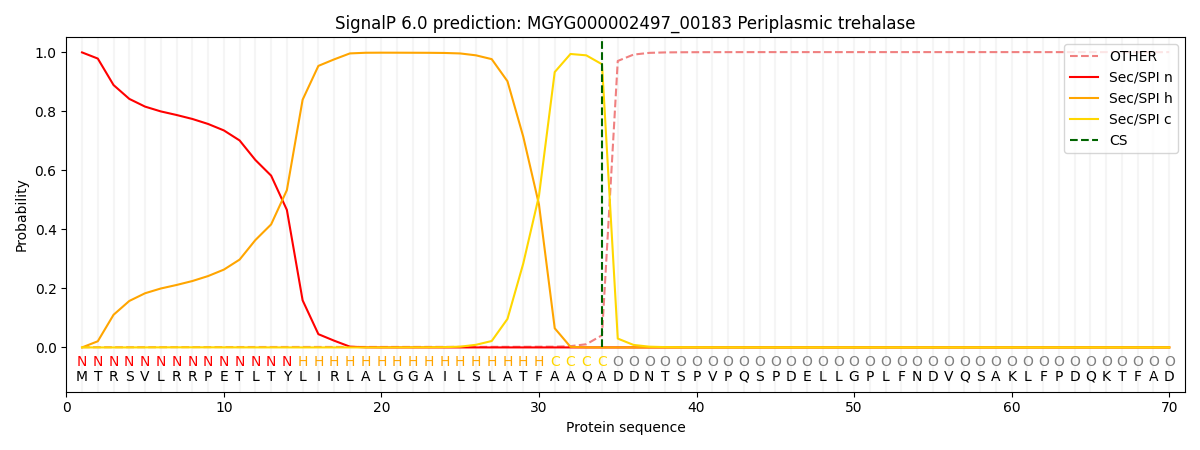
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001650 | 0.997207 | 0.000442 | 0.000293 | 0.000200 | 0.000173 |