You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003205_00074
You are here: Home > Sequence: MGYG000003205_00074
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
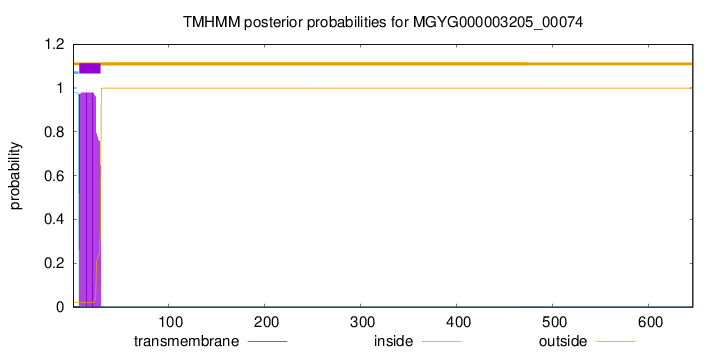
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900760055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900760055 | |||||||||||
| CAZyme ID | MGYG000003205_00074 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 46021; End: 47961 Strand: + | |||||||||||
Full Sequence Download help
| MKIFKRLIVF VSVFSFLAIA LICAENGISE TPQNPYGICA HIAKNEYPVY RKAIDTMEAV | 60 |
| GIGTFRSGAN WINAEKTKGK FDFSRWESLI SYAEKKGIEP LVIFPSNTPS FAKPFPKHID | 120 |
| ELAEVCKRFA GHFKDKMKYF EVVNEPNLKI FWDGEDPDAR AYANLLKRVS TAIKEGNPRA | 180 |
| KVLYSGVAGT PVEFIEESFK AGAGEYFDIM NFHPYSWYDI PEKTLLANIE KVKGLMHKYG | 240 |
| IEDKPVWITE FGYTSAQAHP SIPKYVAKSL KILGVDKAKG KIAYIHDERF AYFGNLFNGS | 300 |
| AEWLFPQAKN FHRINFDEIK NLSPSDCPVL YILPNEIFPK KYRSDLLEYV KKGGTLVTVG | 360 |
| GIPFYYDTEG LENGKAKRKV AGRSPLKAFR MGVKTFKDYG FVEPNLVKMR IHPGRRVAKT | 420 |
| ESAEGFSDIK CEGYFPPYFI STENTTPDDE VIPIVYGVFG DKKLPLGAIY KLGGEFKGNI | 480 |
| IAFYGILGES ASEQMQAKML PRQFILARSR GIERVFKYCL ISDAFDYTRE SHFGIVGKNL | 540 |
| EPKPSYYAYK TLAETLENSL PEYRSCGNIN IAEWVREKDG VPVAAIWSNY YKQPVNIKFE | 600 |
| GKIISAKNYI GEKVGIAAEG QTLSLDADMG VLYLEGVKNV EICKSE | 646 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 74 | 258 | 4e-28 | 0.505800464037123 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01229 | Glyco_hydro_39 | 3.30e-07 | 116 | 273 | 121 | 295 | Glycosyl hydrolases family 39. |
| cd21510 | agarase_cat | 1.02e-04 | 114 | 255 | 71 | 234 | alpha-beta barrel catalytic domain of agarase, such as GH86-like endo-acting agarases identified in non-marine organisms. Typically, agarases (E.C. 3.2.1.81) are found in ocean-dwelling bacteria since agarose is a principle component of red algae cell wall polysaccharides. Agarose is a linear polymer of alternating D-galactose and 3,6-anhydro-L-galactopyranose. Endo-acting agarases, such as glycoside hydrolase 16 (GH16) and GH86 hydrolyze internal beta-1,4 linkages. GH86-like endo-acting agarase of this protein family has been identified in the human intestinal bacterium Bacteroides uniformis. This acquired metabolic pathway, as demonstrated by the prevalence of agar-specific genetic cluster called polysaccharide utilization loci (PULs), varies considerably between human populations, being much more prevalent in a Japanese sample than in North America, European, or Chinese samples. Agarase activity was also identified in the non-marine bacterium Cellvibrio sp. |
| pfam11790 | Glyco_hydro_cc | 1.95e-04 | 144 | 255 | 71 | 173 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
| smart00633 | Glyco_10 | 0.002 | 71 | 160 | 3 | 96 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 0.002 | 71 | 146 | 69 | 151 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44885.1 | 2.46e-102 | 34 | 640 | 27 | 614 |
| QDU57372.1 | 1.54e-68 | 27 | 638 | 21 | 626 |
| AVM44901.1 | 1.29e-43 | 34 | 618 | 25 | 608 |
| AHF90941.1 | 3.01e-30 | 24 | 588 | 212 | 803 |
| ACZ43011.1 | 3.94e-27 | 34 | 255 | 302 | 530 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1PX8_A | 5.12e-06 | 116 | 251 | 124 | 278 | Crystalstructure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1PX8_B Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1UHV_A Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1UHV_B Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1UHV_C Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1UHV_D Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum] |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
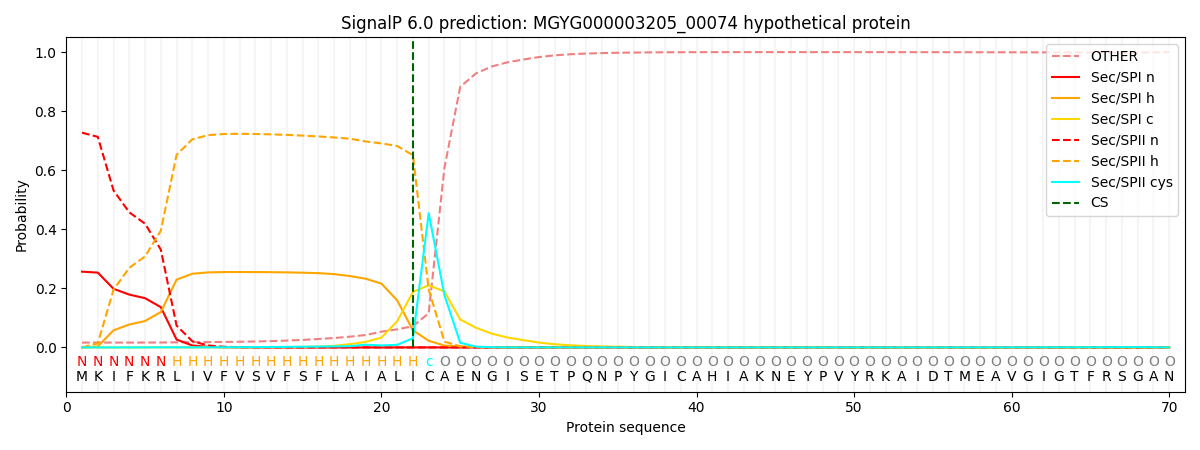
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.018126 | 0.249359 | 0.731936 | 0.000207 | 0.000164 | 0.000197 |