You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001319_00425
You are here: Home > Sequence: MGYG000001319_00425
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
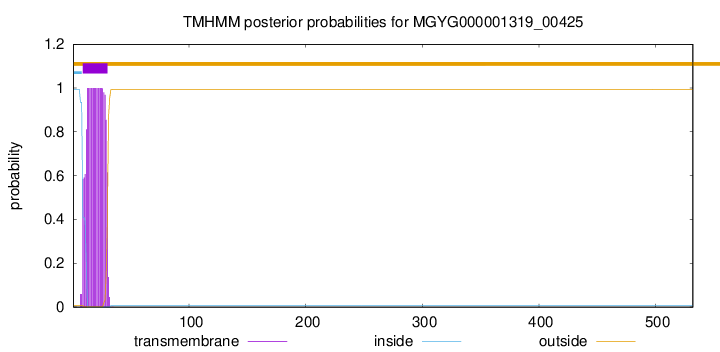
TMHMM annotations
Basic Information help
| Species | Butyrivibrio_A crossotus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Butyrivibrio_A; Butyrivibrio_A crossotus | |||||||||||
| CAZyme ID | MGYG000001319_00425 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 277994; End: 279592 Strand: - | |||||||||||
Full Sequence Download help
| MTEDKRKKII TGCIIAGMVV LIIVLTTVIL KVKKTKVGTA DKTDPAEYSS SDITSKETES | 60 |
| VTVPQEQPTE YSSSDITSKE TESITVPQEQ PTTPAGNDKI TNTVNYSVIL KKNNSWSDGT | 120 |
| NEFFQYECIV TNNTGYDITS WKFEGNIGNV ASIESSWNGN FAYKDGMLVI TPLDWNGNIK | 180 |
| AGQTVSTCGV IIKGSNLKES VEYGSSCNNN YNNNTSASTE TQYELPELEN GTPLANHGKL | 240 |
| SVNGVNLVDS KGQKFQLKGV STHGIQWFPQ YVNKDTFKDL RDNWGANVIR LAMYSAEGGY | 300 |
| CNGNTAEFDK IIDRGVQACS ELGMYVIIDW HILSDSNPNT NKEQAKQFFA KMSKQYADCN | 360 |
| NVIYEICNEP NGGVSWSEIK KYADEVIPVI RQYSKDAIII CGTPTWSQDV DLVASDPLQD | 420 |
| GHNVMYAVHF YAATHKDDLR NKVINAIKAG TPVFVSEFSI CDASGNGGID YDSAGKWKEL | 480 |
| INNYNLSFAG WSLCNKGETS ALIKSGVSRL SGFTTSDLSE TGIWLRNFIS GK | 532 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 251 | 488 | 2e-95 | 0.9831223628691983 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.26e-67 | 248 | 497 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 2.11e-15 | 239 | 501 | 49 | 368 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| smart00637 | CBD_II | 4.81e-13 | 113 | 196 | 1 | 80 | CBD_II domain. |
| pfam00553 | CBM_2 | 6.78e-09 | 103 | 186 | 2 | 77 | Cellulose binding domain. Two tryptophan residues are involved in cellulose binding. Cellulose binding domain found in bacteria. |
| smart01063 | CBM49 | 1.36e-05 | 107 | 195 | 1 | 84 | Carbohydrate binding domain CBM49. This domain is found at the C terminal of cellulases and in vitro binding studies have shown it to binds to crystalline cellulose. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWT53734.1 | 3.78e-151 | 108 | 532 | 75 | 515 |
| QNM00780.1 | 1.44e-145 | 108 | 532 | 75 | 515 |
| CBK83877.1 | 5.45e-138 | 130 | 532 | 102 | 530 |
| QNM02472.1 | 7.38e-118 | 224 | 528 | 78 | 383 |
| ACR73731.1 | 1.38e-116 | 111 | 532 | 73 | 536 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 9.74e-108 | 232 | 529 | 5 | 300 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 3PZT_A | 2.47e-103 | 232 | 531 | 29 | 326 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 4XZW_A | 1.97e-101 | 232 | 526 | 4 | 299 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 4XZB_A | 4.56e-100 | 232 | 526 | 4 | 300 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 1H11_A | 2.78e-92 | 229 | 529 | 1 | 300 | 2-DEOXY-2-FLURO-B-D-CELLOTRIOSYL/ENZYMEINTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.08 ANGSTROM RESOLUTION [Salipaludibacillus agaradhaerens],1H2J_A ENDOGLUCANASE CEL5A IN COMPLEX WITH UNHYDROLYSED AND COVALENTLY LINKED 2,4-DINITROPHENYL-2-DEOXY-2-FLUORO-CELLOBIOSIDE AT 1.15 A RESOLUTION [Salipaludibacillus agaradhaerens],1HF6_A ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE ORTHORHOMBIC CRYSTAL FORM IN COMPLEX WITH CELLOTRIOSE [Salipaludibacillus agaradhaerens],1OCQ_A COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.08 ANGSTROM RESOLUTION with cellobio-derived isofagomine [Salipaludibacillus agaradhaerens],1W3K_A Endoglucanase Cel5a From Bacillus Agaradhaerens In Complex With Cellobio Derived-tetrahydrooxazine [Salipaludibacillus agaradhaerens],1W3L_A Endoglucanase Cel5a From Bacillus Agaradhaerens In Complex With Cellotri Derived-Tetrahydrooxazine [Salipaludibacillus agaradhaerens],4A3H_A 2',4' Dinitrophenyl-2-Deoxy-2-Fluro-B-D-Cellobioside Complex Of The Endoglucanase Cel5a From Bacillus Agaradhaerens At 1.6 A Resolution [Salipaludibacillus agaradhaerens],5A3H_A 2-Deoxy-2-Fluro-B-D-CellobiosylENZYME INTERMEDIATE COMPLEX Of The Endoglucanase Cel5a From Bacillus Agaradhearans At 1.8 Angstroms Resolution [Salipaludibacillus agaradhaerens],6A3H_A 2-Deoxy-2-Fluro-B-D-CellotriosylENZYME INTERMEDIATE COMPLEX OF THE Endoglucanase Cel5a From Bacillus Agaradhearans At 1.6 Angstrom Resolution [Salipaludibacillus agaradhaerens],7A3H_A Native Endoglucanase Cel5a Catalytic Core Domain At 0.95 Angstroms Resolution [Salipaludibacillus agaradhaerens],8A3H_A Cellobiose-derived imidazole complex of the endoglucanase cel5A from Bacillus agaradhaerens at 0.97 A resolution [Salipaludibacillus agaradhaerens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P15704 | 6.26e-101 | 238 | 532 | 45 | 338 | Endoglucanase OS=Clostridium saccharobutylicum OX=169679 GN=eglA PE=3 SV=1 |
| P10475 | 2.84e-100 | 232 | 531 | 34 | 331 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P07983 | 7.95e-100 | 232 | 531 | 34 | 331 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P23549 | 2.71e-97 | 232 | 531 | 34 | 331 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
| P22541 | 6.60e-95 | 219 | 530 | 93 | 408 | Endoglucanase A OS=Butyrivibrio fibrisolvens OX=831 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
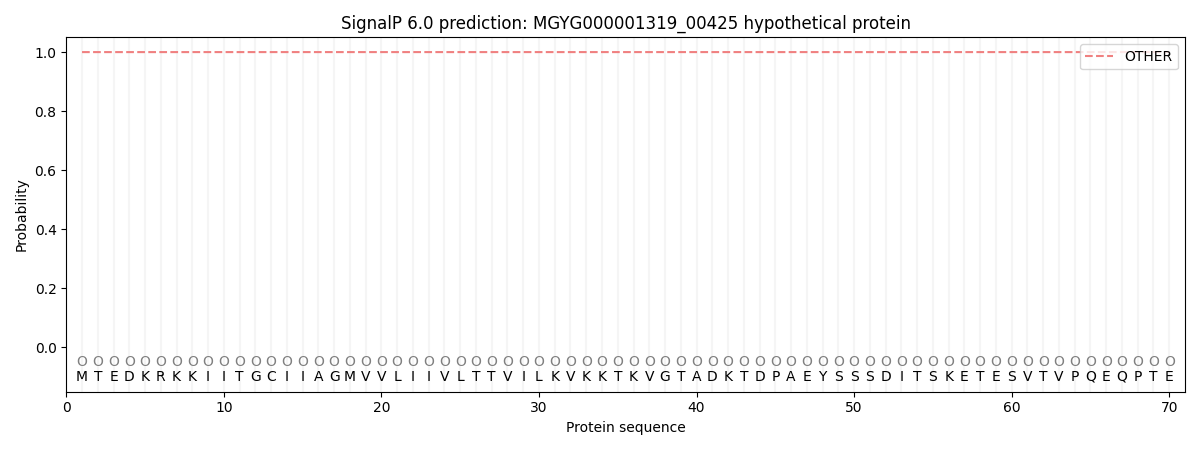
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999991 | 0.000040 | 0.000001 | 0.000000 | 0.000000 | 0.000003 |