You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001645_00071
You are here: Home > Sequence: MGYG000001645_00071
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
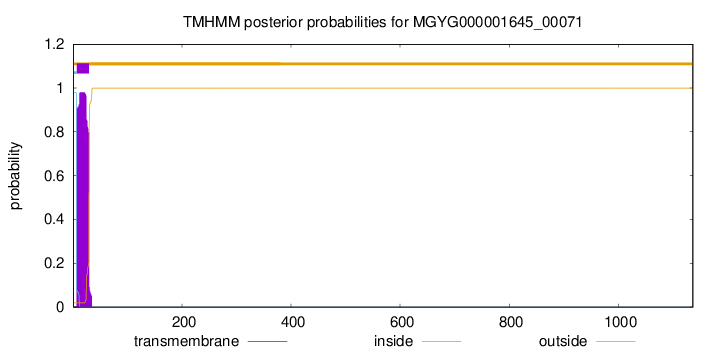
TMHMM annotations
Basic Information help
| Species | Eubacterium_G sp900550135 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_G; Eubacterium_G sp900550135 | |||||||||||
| CAZyme ID | MGYG000001645_00071 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30131; End: 33544 Strand: + | |||||||||||
Full Sequence Download help
| MKKVRKLVSM LLVFAMTFSV AISGKITGIT QVSAREALGS NDFLKVNGTQ IRKQKGTGDV | 60 |
| VYLRGTNAGG WLVQEDWMNP TNASDQKTMM TTLANRFGTS KRDELVSTYE NNYWTTQDFD | 120 |
| NCAEMGMSVI RLPFTYMNLC DDNGNLKSNA FDRLDWFVQN CSQRGMYVIL DMHGAFSSQN | 180 |
| GMDHSGEIND GKQLYYNQNN KDKTLNLWKK IAEHFKGNPA VAAYDILNEP GIKAAATYSL | 240 |
| HWDFYNEIYN TIRSKDSNHI IIMESCWDAD NLPRPSQYGW TNVAYEYHYY PWNAQNSSDA | 300 |
| QKSYFSGKVS DIANHNYGVP TFVGEFTCFE QEEGWKAAMS TFNGQGWHWT TWSYKVTGNS | 360 |
| SWGIYNHNPE KVDIYNDSAD TIKNKWSAVG TANGWKNDKI YNLVKAYLPG TVTSTGGSTT | 420 |
| DDSDNNTTSG VATFYEHSNY GGWTVSLEEG SYDYKDILAK GIVNDQISSL RVSDGYKVTI | 480 |
| YDDEGFKGTS KEFTSDASNV GDEMNDKTSS IKIEKINNQT STTTSYDTVK LPDGKYSIKS | 540 |
| VANEKYVVAE NGGSDPIVAN RDSYGGSWET FYIVNNDDGT VSIKADANNK YVCAVLDEEN | 600 |
| QLTPRSDSIG TWEKFKIYKI NDSEYGIRSA ENRKYVKADL DNGGKLIAGS DSIAGAWEAF | 660 |
| NIEKVGDTTT NNNVATFYEN SNYGGWSVSL PEGTYDYSDI IAKGIKNDAI SSLKVNSGYK | 720 |
| VTLYDNAGFK GTSKAFTGDA SYVGDEMNDK TSSIKIEKWD GSSNVTYNTV KLSNGKYSIK | 780 |
| SVANEKYVVA ENGGSETIVA NRDSYGGSWE TFYLINNDDG TVSIKADANN KYVCAVLDEE | 840 |
| NQLVPRSDNV GAWEKFQIYK ISDTEYGLRS AENGKYVKAD LDNGGKLIAG SDSIAGAWEA | 900 |
| FNIEKLGDET SSAKATFYEN SNYSGWSVAL SEGRYDYGTM ISKGIKNDQI SSVKVADGYK | 960 |
| VTLYNDAGFA GSKKTLFTDA SGLGDFNDKA SAIVIEKVEK ADFNNSNAYI TSIANGKVVC | 1020 |
| AENGGSETIV ANRSSCGGAW ETFQIVNNND GTVSLKSIAN GKYVCAVIDE NNQLLPRSES | 1080 |
| VGTWEKFIIE KISDGEYALY SLANGKYVQA NLNDGGKLFA TSETVAGAWE VFDINRN | 1137 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 92 | 355 | 3.5e-115 | 0.9961977186311787 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.83e-25 | 111 | 354 | 21 | 268 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 2.86e-18 | 61 | 386 | 16 | 381 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd00257 | Fascin | 2.38e-11 | 1015 | 1134 | 10 | 119 | Fascin-like domain; members include actin-bundling/crosslinking proteins facsin, histoactophilin and singed; identified in sea urchin, Drosophila, Xenopus, rodents, and humans; The fascin-like domain adopts a beta-trefoil topology and contains an internal threefold repeat; the fascin subgroup contains four copies of the domain; Structurally similar to fibroblast growth factor (FGF) |
| cd00257 | Fascin | 1.10e-08 | 784 | 901 | 10 | 117 | Fascin-like domain; members include actin-bundling/crosslinking proteins facsin, histoactophilin and singed; identified in sea urchin, Drosophila, Xenopus, rodents, and humans; The fascin-like domain adopts a beta-trefoil topology and contains an internal threefold repeat; the fascin subgroup contains four copies of the domain; Structurally similar to fibroblast growth factor (FGF) |
| smart00247 | XTALbg | 1.65e-05 | 429 | 471 | 40 | 80 | Beta/gamma crystallins. Beta/gamma crystallins |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZS15177.1 | 1.62e-204 | 32 | 668 | 37 | 565 |
| VEI36204.1 | 6.33e-183 | 36 | 665 | 38 | 550 |
| AIC93076.1 | 1.38e-118 | 39 | 407 | 26 | 396 |
| BCJ97694.1 | 2.60e-115 | 31 | 609 | 35 | 522 |
| QAA34430.1 | 9.12e-105 | 23 | 407 | 14 | 405 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3SO1_A | 6.31e-18 | 676 | 756 | 7 | 87 | Crystalstructure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_B Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_C Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_D Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_E Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_F Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_G Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_H Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
| 3SO0_A | 1.61e-17 | 676 | 756 | 7 | 87 | Crystalstructure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_B Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_C Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_D Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_E Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_F Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_G Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_H Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
| 3SNZ_A | 2.99e-17 | 676 | 756 | 7 | 87 | Crystalstructure of a mutant W39D of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
| 3IAJ_A | 3.07e-17 | 676 | 756 | 4 | 84 | Crystalstructure of a betagamma-crystallin domain from Clostridium beijerinckii-in alternate space group I422 [Clostridium beijerinckii NCIMB 8052] |
| 3I9H_A | 3.16e-17 | 676 | 756 | 5 | 85 | Crystalstructure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_B Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_C Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_D Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_E Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_F Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_G Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_H Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| W8QRE4 | 1.36e-24 | 43 | 290 | 3 | 248 | Beta-xylosidase OS=Phanerodontia chrysosporium OX=2822231 GN=Xyl5 PE=1 SV=2 |
| B0XN12 | 1.45e-19 | 63 | 263 | 42 | 245 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=exgA PE=3 SV=1 |
| Q4WK60 | 1.93e-19 | 63 | 256 | 42 | 238 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=exgA PE=3 SV=1 |
| A1D4Q5 | 6.18e-19 | 63 | 253 | 42 | 235 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=exgA PE=3 SV=1 |
| Q7Z9L3 | 1.32e-18 | 63 | 262 | 31 | 232 | Glucan 1,3-beta-glucosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=exgA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
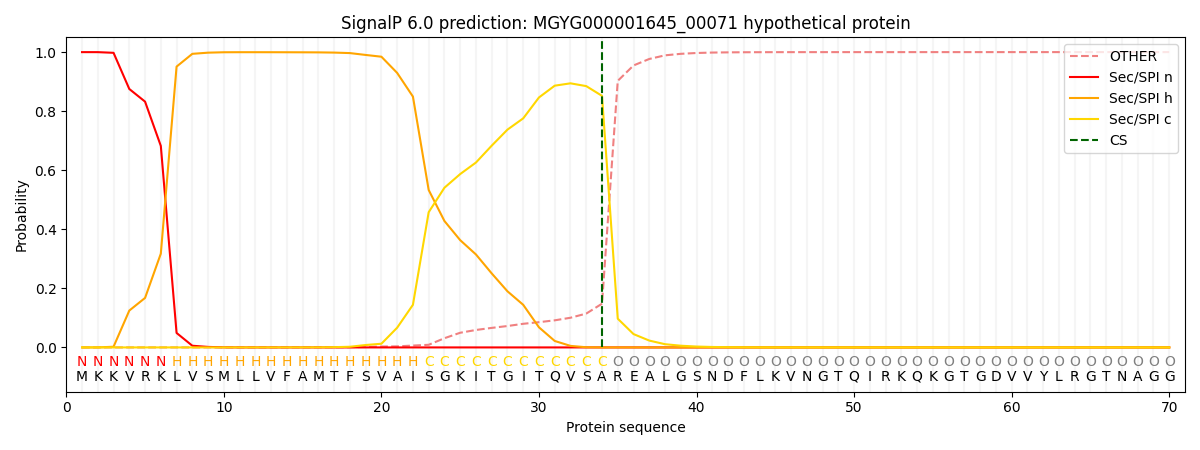
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000332 | 0.998865 | 0.000316 | 0.000161 | 0.000147 | 0.000145 |