You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002321_00026
You are here: Home > Sequence: MGYG000002321_00026
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus_D bicirculans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_D; Ruminococcus_D bicirculans | |||||||||||
| CAZyme ID | MGYG000002321_00026 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30731; End: 32365 Strand: - | |||||||||||
Full Sequence Download help
| MVKLISLKKA AAVLLTSVTL MSAAAALPAV DETPLLSQTA ITVEAASVGK VTGLTSKTLS | 60 |
| NSEIKLSWKK VSGASGYSVC MRKNGKYPQI ADVKSGSTLT YTVKNLPNAT RENFKVRAYK | 120 |
| TVKGKKVYGA YSDNWNTATN PQPAKGLKVS SVSYDSVKLS WTKIGCTNYR VFQLKNGQWK | 180 |
| EIAKTTGTSY TVKNLSQKTT YKFKIRACKT DDKKANHYGK YSAEVSATTS KAPAVVTPVS | 240 |
| QHGQLSVKGA NIVDKNGKVF KIKGMSTHGI MWEDFSDILT KDSLKVLRDD WKVNTIRIAM | 300 |
| YTEEWGGYCT ENGKYQAQAK QKVKTGVENA KSLGMYAIID WHVLSDQNPN NHKNDAIKFF | 360 |
| TEMAKTYKDY NNVIYEICNE PNGGVTWTGG IKSYCQSVVS TIRKYDSDAI IICGTGTWSQ | 420 |
| DIDQVLGNKL SDKNCVYALH FYANTHTDWL RNRLQNCYKK GLPVLVSEFG TCDASGNGGY | 480 |
| NSTESTKWLK LLDSLKVGYI NWSACGKSET ASAFNSGTNL KAIKSGTSQL TASGKFIRDW | 540 |
| YRNH | 544 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 255 | 501 | 1.2e-89 | 0.9957805907172996 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.20e-68 | 253 | 508 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 3.99e-14 | 251 | 511 | 46 | 369 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd00063 | FN3 | 9.00e-11 | 141 | 229 | 1 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| smart00060 | FN3 | 6.93e-09 | 141 | 207 | 1 | 76 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
| pfam00041 | fn3 | 3.54e-05 | 143 | 207 | 2 | 75 | Fibronectin type III domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCO03766.1 | 0.0 | 1 | 544 | 1 | 544 |
| CBL33590.1 | 4.78e-101 | 233 | 544 | 279 | 585 |
| CBK95395.1 | 1.98e-99 | 226 | 544 | 255 | 571 |
| QHZ49094.1 | 2.59e-98 | 234 | 543 | 30 | 330 |
| QNM02472.1 | 3.44e-98 | 230 | 543 | 77 | 386 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 3.18e-103 | 237 | 542 | 5 | 301 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 3PZT_A | 3.14e-98 | 230 | 539 | 22 | 322 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 4XZW_A | 2.22e-97 | 237 | 538 | 4 | 299 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 4XZB_A | 4.51e-94 | 237 | 538 | 4 | 300 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 1A3H_A | 2.52e-82 | 239 | 542 | 3 | 298 | EndoglucanaseCel5a From Bacillus Agaradherans At 1.6a Resolution [Salipaludibacillus agaradhaerens],2A3H_A Cellobiose Complex Of The Endoglucanase Cel5a From Bacillus Agaradherans At 2.0 A Resolution [Salipaludibacillus agaradhaerens],3A3H_A Cellotriose Complex Of The Endoglucanase Cel5a From Bacillus Agaradherans At 1.6 A Resolution [Salipaludibacillus agaradhaerens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10475 | 8.43e-96 | 227 | 539 | 24 | 327 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P07983 | 6.58e-95 | 230 | 539 | 27 | 327 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P23549 | 7.91e-93 | 227 | 539 | 24 | 327 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
| Q59394 | 1.39e-91 | 237 | 542 | 33 | 329 | Endoglucanase N OS=Pectobacterium atrosepticum OX=29471 GN=celN PE=3 SV=1 |
| Q47096 | 7.95e-91 | 237 | 542 | 33 | 329 | Endoglucanase 5 OS=Pectobacterium carotovorum subsp. carotovorum OX=555 GN=celV PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
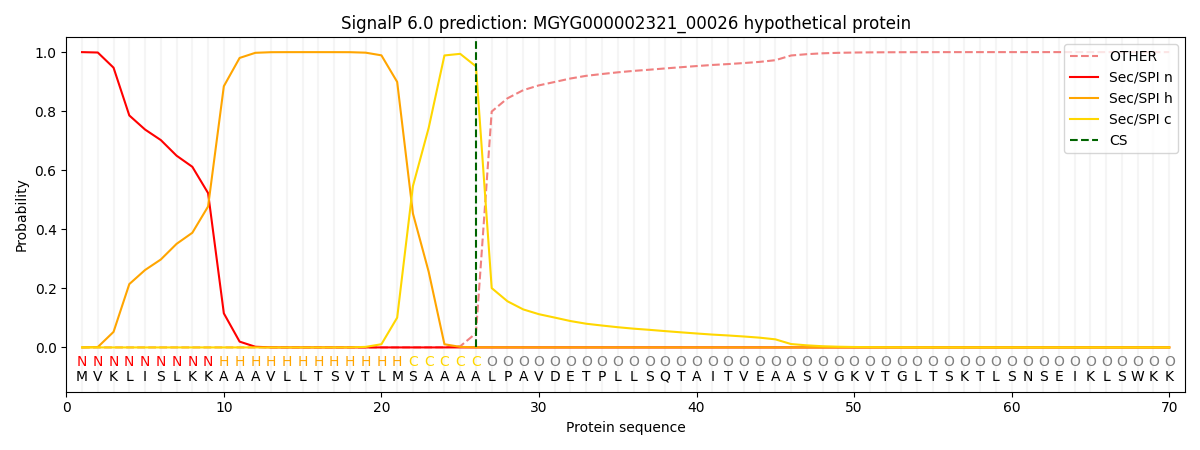
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000530 | 0.998661 | 0.000182 | 0.000230 | 0.000186 | 0.000172 |
