You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002321_00082
You are here: Home > Sequence: MGYG000002321_00082
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus_D bicirculans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_D; Ruminococcus_D bicirculans | |||||||||||
| CAZyme ID | MGYG000002321_00082 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 99634; End: 100842 Strand: + | |||||||||||
Full Sequence Download help
| MKVKQWLASS LAVLMLVSTV GCGDNSSSSK AESKSNNSSA VVSETAMPEK DYATTTDGSA | 60 |
| KVKIDGTKFM VGDKELWFNG VNAPWDKWND FGGGFNFEFW QDHFQKLHNS GVNAARIWII | 120 |
| CNGDVGMAIS ADGTFDGATT AHWEDLDNLF YLAEQYQIYI MATVQSFDNF KDQNQNYQAW | 180 |
| RTLIQDSDKT DMFVDNYIVP LVQRYGKSDY FWSVDLCNEP DWIVENEECG KLDWLYLEQY | 240 |
| YAKAAAAIHA NSDVLVTVGM GMIKYNSDSQ QGNKISDSAL QDVLSGDKYD KSLAYVDFYS | 300 |
| THWYTWMQGM WGYPFSESPT DFGLDGTKPC VIGECPAVAS DSDFDITSAY EDAYNNGWNG | 360 |
| VFAWKTSGQD DGCGLWLDIQ PAIEKMAGIC EDKIFPNGKK AV | 402 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 74 | 366 | 6e-41 | 0.9836065573770492 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCO03823.1 | 5.32e-311 | 1 | 402 | 1 | 402 |
| ADD61974.1 | 4.84e-282 | 1 | 402 | 1 | 411 |
| ADU21807.1 | 5.34e-191 | 24 | 400 | 35 | 411 |
| CBL16471.1 | 2.58e-129 | 61 | 399 | 32 | 364 |
| QHT63496.1 | 1.38e-112 | 46 | 397 | 23 | 367 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3VUP_A | 9.58e-20 | 61 | 364 | 1 | 322 | Beta-1,4-mannanasefrom the common sea hare Aplysia kurodai [Aplysia kurodai],3VUP_B Beta-1,4-mannanase from the common sea hare Aplysia kurodai [Aplysia kurodai] |
| 4OOU_A | 2.52e-19 | 53 | 371 | 14 | 348 | Crystalstructure of beta-1,4-D-mannanase from Cryptopygus antarcticus [Cryptopygus antarcticus],4OOU_B Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus [Cryptopygus antarcticus],4OOZ_A Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus in complex with mannopentaose [Cryptopygus antarcticus],4OOZ_B Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus in complex with mannopentaose [Cryptopygus antarcticus] |
| 5Y6T_A | 1.93e-15 | 64 | 371 | 10 | 333 | ChainA, endo-1,4-beta-mannanase [Eisenia fetida] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B4XC07 | 1.31e-18 | 53 | 371 | 14 | 348 | Mannan endo-1,4-beta-mannosidase OS=Cryptopygus antarcticus OX=187623 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
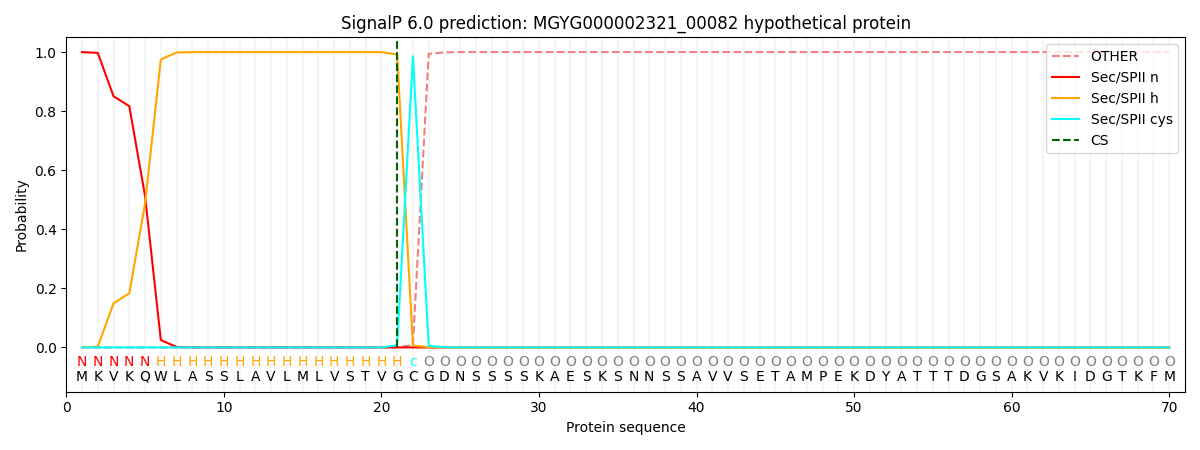
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000013 | 1.000045 | 0.000000 | 0.000000 | 0.000000 |
