You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003640_00313
You are here: Home > Sequence: MGYG000003640_00313
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
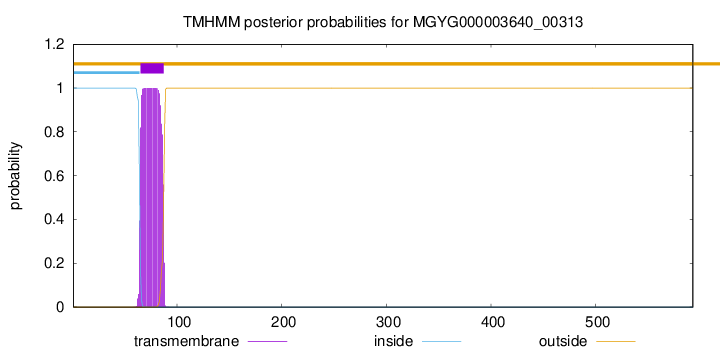
TMHMM annotations
Basic Information help
| Species | Butyrivibrio_A sp900771195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Butyrivibrio_A; Butyrivibrio_A sp900771195 | |||||||||||
| CAZyme ID | MGYG000003640_00313 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5475; End: 7256 Strand: - | |||||||||||
Full Sequence Download help
| MSDQINDESK AENLSAENPE QNSAELGMTE NPEENSAEPD MTENQKEDSD ASLPKEKKHN | 60 |
| RYKVLCVILA ACLFVAMVFT GLFATGVLKP FGSQAEPMES TQSVTVPETK TEVTEAEETD | 120 |
| AEATEEADAT EAGTEEETAG GEDDAGTDAK TATFTVDFVV ANSWGSGSAT NYQYSVSVRN | 180 |
| TSRVDIKTWG QIEADVPEGT SVTQAWGCEY ELKDNKLILK PTEYTAEIAA GAVRGDIGLI | 240 |
| LESPGKMENF SCDGTTDSVA SSSGSSDGGS SYPDPSTPVT PYEPPELEAG TPVGNHGQLS | 300 |
| VKGTDLVDQN GKKFQLKGVS THGIGWFPDY VNEDGFKTLR DNWGANLIRI AMYTDEYNGY | 360 |
| CSGGNKEELE ALVCKGVEAC TDLGMYVIID WHVLHDLNPN KNIDSAKVFF DRMSQKYAGN | 420 |
| VNVIYEICNE PNGDTTWADI KKYADVIIPI IRKNSPNAVI IVGTPTWSQD VDQVAANPLK | 480 |
| NGENVMYALH FYAATHKDDL RNKLTKARED GTPIFVSEFS ICDASGNGGI DYDSAEKWKE | 540 |
| LINSENLSYA GWSLSNKAET SALINPSCTK LSGWESGDLS STGLWLRDMI SGK | 593 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 309 | 550 | 1.4e-93 | 0.9915611814345991 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.74e-71 | 307 | 557 | 1 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 9.58e-18 | 291 | 496 | 39 | 227 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| smart00637 | CBD_II | 6.72e-07 | 162 | 232 | 2 | 67 | CBD_II domain. |
| COG3867 | GanB | 0.005 | 292 | 392 | 31 | 125 | Arabinogalactan endo-1,4-beta-galactosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWT53734.1 | 1.89e-159 | 161 | 593 | 80 | 515 |
| QNM00780.1 | 8.63e-158 | 137 | 593 | 54 | 515 |
| CBK83877.1 | 4.82e-147 | 137 | 593 | 62 | 530 |
| ADD61840.1 | 1.53e-127 | 63 | 593 | 5 | 539 |
| ACR73731.1 | 1.97e-127 | 63 | 593 | 5 | 536 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 1.88e-108 | 291 | 590 | 5 | 300 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 3PZT_A | 3.70e-103 | 291 | 592 | 29 | 326 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 4XZW_A | 3.59e-99 | 291 | 587 | 4 | 299 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 4XZB_A | 5.09e-96 | 291 | 587 | 4 | 300 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 1A3H_A | 9.64e-92 | 293 | 590 | 3 | 297 | EndoglucanaseCel5a From Bacillus Agaradherans At 1.6a Resolution [Salipaludibacillus agaradhaerens],2A3H_A Cellobiose Complex Of The Endoglucanase Cel5a From Bacillus Agaradherans At 2.0 A Resolution [Salipaludibacillus agaradhaerens],3A3H_A Cellotriose Complex Of The Endoglucanase Cel5a From Bacillus Agaradherans At 1.6 A Resolution [Salipaludibacillus agaradhaerens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q07940 | 6.90e-102 | 312 | 583 | 17 | 287 | Endoglucanase 4 OS=Ruminococcus albus OX=1264 GN=Eg IV PE=1 SV=1 |
| P07983 | 1.53e-100 | 285 | 592 | 26 | 331 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P22541 | 2.93e-100 | 296 | 591 | 113 | 408 | Endoglucanase A OS=Butyrivibrio fibrisolvens OX=831 GN=celA PE=1 SV=1 |
| P10475 | 4.28e-100 | 285 | 592 | 26 | 331 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P23549 | 1.02e-97 | 285 | 592 | 26 | 331 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
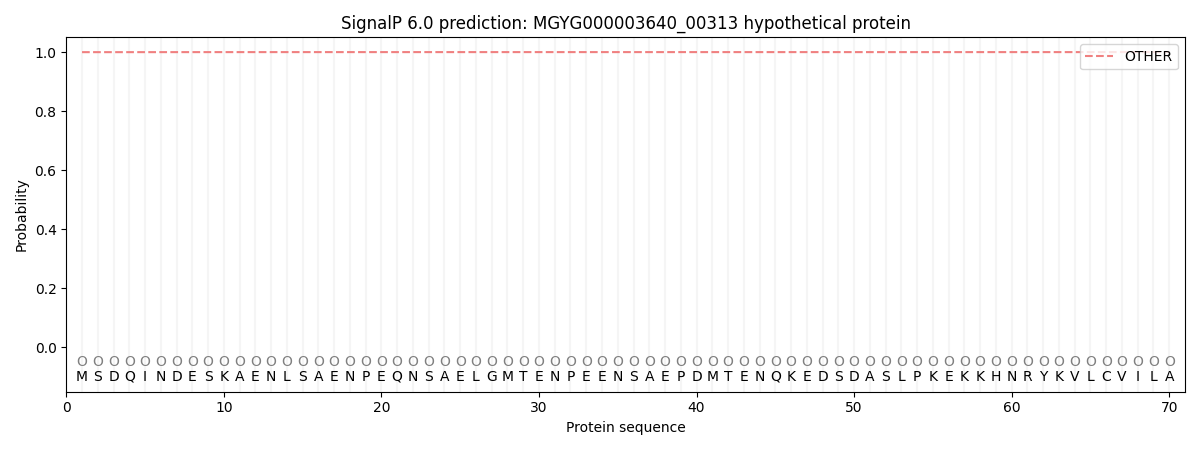
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999866 | 0.000100 | 0.000015 | 0.000000 | 0.000000 | 0.000011 |