You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001018_00245
You are here: Home > Sequence: MGYG000001018_00245
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
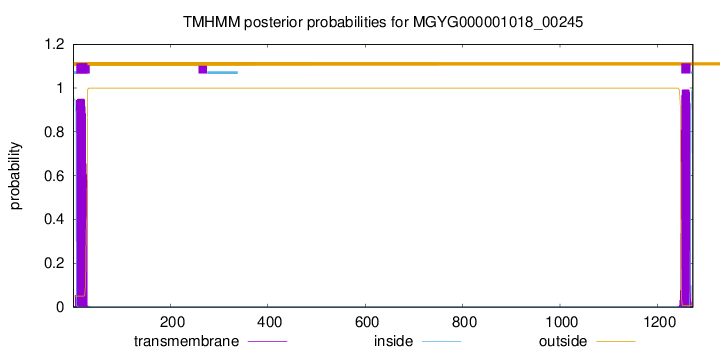
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter sp900751785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter sp900751785 | |||||||||||
| CAZyme ID | MGYG000001018_00245 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6392; End: 10213 Strand: - | |||||||||||
Full Sequence Download help
| MKVRRVFLAL ILSVSCAFSG SFAALAQAAE GQAVALDEGL RGYWDFEGED PRENKGSDAS | 60 |
| LSGSLSGNAV SVQQSSVEEM GSVLHFDTRS GESSRMLIAS ALNSGGEDFS ISLWVNHSSQ | 120 |
| QNASGKTILL QQSGSGRTLL YRQNGQYVTY ISAADVAMGT STGSDIWEHI VLVRSGEPSA | 180 |
| YQLTLYVNGE KANERNLTAG AVDAVTDLII GAHKNAGDIG QFVGDIDELR LYDRAVTAEE | 240 |
| AEALYEEHED VVLEQQLQGI KKELKALVDE ACEVYENGPL DKEDPAAAAL AAAISGAELA | 300 |
| LEGIDADALR NASEALENAL NAYMDGSLIL TIDTENVSRT VDDGIFGINH RYAFNGYGSF | 360 |
| DSETMQIKEE FAELYSQAGF GSLRYPGGTI SNLFNWKDTI GPKEERVAQI HGFYNNSGQH | 420 |
| GIAPNFGVDE AAEFVQEYDS ELVYVYALAR GDADDAADLI EYLNAEVGSN PNGGTAWAEV | 480 |
| RAANGHEEPY NVRCFEIGNE MNQGGADGTT AQTYWIDDVP GGALEGYVNG GTASFTRQYA | 540 |
| VALDDWNQSA SYSDGSADQV FYLRYARVER DENADDYGSF TAVNPGSVKV YVGGEAWTET | 600 |
| DDLSSAGAED KVYAVDYKTG AISFGDGEHG AVPASGSQIT ADYSVDRDGF VQISEAMRST | 660 |
| MDQINEYNAQ NGLAEKEIHI YSSYETENFV QKMHNEGHDD LYDGLTIHPY SGTPSGGSST | 720 |
| EESRKSFYYD AMLKGDAKAA DVENYVKLMQ QYDETKVPVI SEYGIFRSTD TMVRSQTHAL | 780 |
| YIARAIMEYV RLGSPYIQKH CLVDWYSDGA DSLGPTQQAV IQAVSTGGST ADGEGGFTFF | 840 |
| ATPSARVFEM LNTAFGDQIT ASSLNYTQQL DNGVEQYSVM TSKDDEGNVY AAIVNLNLDR | 900 |
| ENKIKIQVDD LDLTGKTMEI QEISGDTFYA ENTPDDPDNV KVERSSAAIE GTAAEVTLAP | 960 |
| HSFTVVKIVD ALSAEEPEEP EQPLRTAVLE FTLELADSKS TEGVIPSVAE IFEQAKADAR | 1020 |
| AVLDAVAEGD ESVTQEQIDE CWRALMESMQ YLSFLQGDKS DLEKVIEFAA DAEANLDSYV | 1080 |
| ETGKAEFIQV LEEARAVYSD ADAMQEEVDS AWMTLLEKTA DLRLKADKWA LEDLINEAAS | 1140 |
| MDLSAYTEES TAAFHAAFAN ARSVLSDDTL SEDDQHVVDE AAAQLAAARD ALIVKDDAEE | 1200 |
| AAEPTEPADP SGSGGAQKSG TAGENSSNNE EQPGSKAGNS VPVTGDRSVW AVFMVLCLVS | 1260 |
| GVCAVYLSRR RNN | 1273 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3534 | AbfA | 2.24e-22 | 334 | 968 | 26 | 498 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| pfam13385 | Laminin_G_3 | 1.30e-16 | 97 | 237 | 7 | 146 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| TIGR02243 | TIGR02243 | 2.76e-11 | 553 | 645 | 358 | 442 | putative baseplate assembly protein. This family consists of a large, conserved hypothetical protein in phage tail-like regions of at least six bacterial genomes: Gloeobacter violaceus PCC 7421, Geobacter sulfurreducens PCA, Streptomyces coelicolor A3(2), Streptomyces avermitilis MA-4680, Mesorhizobium loti, and Myxococcus xanthus. The C-terminal region is identified by the broader model pfam04865 as related to baseplate protein J from phage P2, but that relationship is not observed directly. [Mobile and extrachromosomal element functions, Prophage functions] |
| smart00813 | Alpha-L-AF_C | 2.93e-10 | 842 | 962 | 67 | 189 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam06964 | Alpha-L-AF_C | 1.69e-09 | 842 | 962 | 70 | 192 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXM90856.1 | 2.61e-272 | 33 | 968 | 45 | 989 |
| ALE10461.1 | 6.10e-272 | 33 | 968 | 28 | 972 |
| BBA48000.1 | 1.35e-270 | 33 | 968 | 28 | 972 |
| BBA56144.1 | 1.91e-270 | 33 | 968 | 28 | 972 |
| AFL03619.1 | 3.79e-270 | 33 | 968 | 28 | 972 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ATW_A | 1.98e-15 | 334 | 501 | 25 | 173 | Thecrystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_B The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_C The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_D The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_E The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_F The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8] |
| 3S2C_A | 2.00e-15 | 334 | 501 | 25 | 173 | Structureof the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_B Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_C Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_D Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_E Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_F Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_G Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_H Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_I Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_J Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_K Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_L Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1] |
| 3UG3_A | 2.18e-15 | 334 | 501 | 45 | 193 | Crystalstructure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG4_A Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG5_A Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima] |
| 2Y2W_A | 2.52e-14 | 335 | 502 | 69 | 217 | Elucidationof the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_B Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_C Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_D Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_E Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_F Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum] |
| 5O7Z_A | 1.89e-13 | 330 | 502 | 5 | 174 | ChainA, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O7Z_B Chain B, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O7Z_C Chain C, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O7Z_D Chain D, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O7Z_E Chain E, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O7Z_F Chain F, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O80_A Chain A, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O80_B Chain B, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O80_C Chain C, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O80_D Chain D, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O80_E Chain E, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O80_F Chain F, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E7CY70 | 3.91e-14 | 335 | 502 | 39 | 188 | Exo-alpha-(1->6)-L-arabinofuranosidase OS=Bifidobacterium longum OX=216816 GN=afuB PE=1 SV=1 |
| Q841V6 | 1.35e-13 | 335 | 502 | 69 | 217 | Intracellular exo-alpha-(1->5)-L-arabinofuranosidase OS=Bifidobacterium longum OX=216816 GN=abfB PE=1 SV=1 |
| A3DIH0 | 1.95e-13 | 330 | 502 | 6 | 175 | Intracellular exo-alpha-(1->5)-L-arabinofuranosidase OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=Cthe_2548 PE=1 SV=1 |
| Q9KBR4 | 2.38e-12 | 335 | 502 | 28 | 176 | Intracellular exo-alpha-(1->5)-L-arabinofuranosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=abfA PE=3 SV=1 |
| P94531 | 9.56e-12 | 335 | 502 | 27 | 175 | Intracellular exo-alpha-(1->5)-L-arabinofuranosidase 1 OS=Bacillus subtilis (strain 168) OX=224308 GN=abfA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
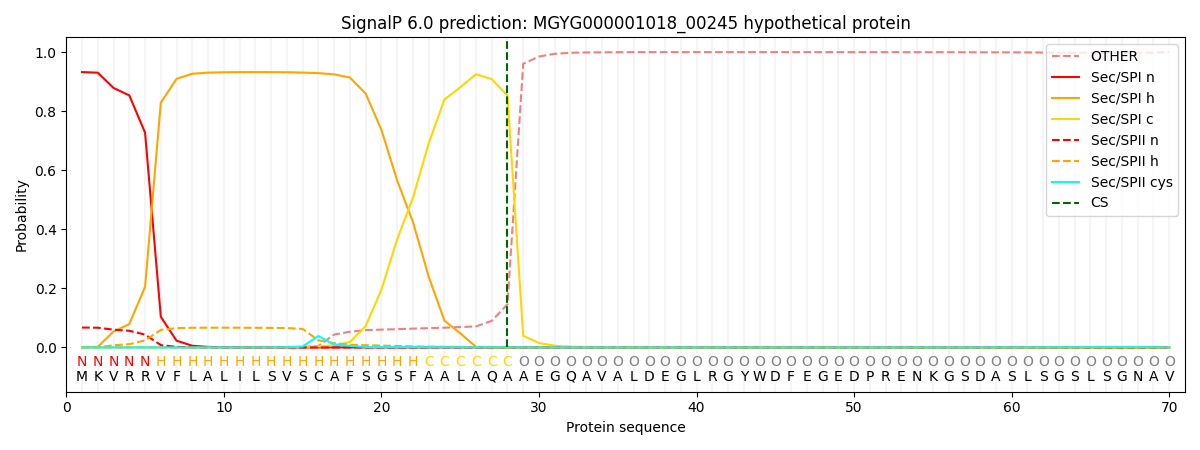
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001242 | 0.924379 | 0.072617 | 0.000993 | 0.000399 | 0.000330 |