You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001360_00035
You are here: Home > Sequence: MGYG000001360_00035
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella salivae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella salivae | |||||||||||
| CAZyme ID | MGYG000001360_00035 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 32980; End: 35568 Strand: + | |||||||||||
Full Sequence Download help
| MKHKIILFLA LCLGGGAVQA QNITPAYTIM GKDSTCQVFV YSPSAREGLH LAFLGDDDQW | 60 |
| HEVGQLCASD YGPWGAEKRM FDPYVTKAAD GTWRAVWSVN DSAPVFAAAY SDDLITWRPQ | 120 |
| DYPLVREKGV KSPVVYQMGD GSFDIYLKTS KGKRYVHATE DFRHFEEDSI PSQADEVLWQ | 180 |
| KDNAVINGKK FLGNAFDVSA YHLQFIRSWF AGLKADNVRY AEQMKDDKSR FATLNRPVEA | 240 |
| TLHVDLTKTK PISDKLIGVF FEDISRAADG GLYAQLLENG DFEYSPADHR GWNSQTAWTS | 300 |
| DKPIVIATAE PLSKNNTHYA VLNQATLTNH GWDNVIYDHG EQFDFSVYAR CIGQKKATLL | 360 |
| VQLVDSLGQV LTEGKVKVEG NSWQPYSLSL NTLTKKRAQP VTPVRCSLRI VSTKPGDVAI | 420 |
| DMVSLFPHDT YKGHGMRKDI AETIAALRPK FMRFPGGCML HGDGINNIYH WKESIGPLYD | 480 |
| RKPDRNIWGY HQTKGLGFFE YFQLCEDLGA APLPVLAAGV PCQNSAANKE GLAGQQCGIP | 540 |
| MNQMPAYVQD VLDLIEWANG NPASSKWAKM RADAGHPAPF HLKMIGIGNE DLITTAFEER | 600 |
| YLMICKAVKA KYPDIEVIGT VGPFHYPSAD YIEGWKVAKA NKSVIDAVDE HYYESPGWFL | 660 |
| HNQNYYDQYD RKGPKVYLGE YASRSRTVES ALAEAIHLCN IERNGDIVEM TSYAPLLCHE | 720 |
| KHQNWNPDLI YFNASQITTT PNYHTQALFS QFSGNQYVTS HVDIAKDLAY RVAVSVVKDS | 780 |
| RTGETMLKLV NALPATINLK LDGISLPGDS RVVGFDGNPS DMKVKTFDAM RQEKLINVKN | 840 |
| SVLQLPAYSV MAISFNSNMV TK | 862 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH51 | 379 | 830 | 5.4e-97 | 0.6936507936507936 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3534 | AbfA | 3.12e-30 | 419 | 853 | 32 | 496 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| pfam06964 | Alpha-L-AF_C | 1.03e-27 | 687 | 849 | 23 | 192 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| smart00813 | Alpha-L-AF_C | 1.21e-21 | 687 | 849 | 19 | 189 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEH15278.1 | 0.0 | 1 | 854 | 1 | 854 |
| BCS84266.1 | 0.0 | 1 | 853 | 21 | 875 |
| AGB28163.1 | 0.0 | 1 | 853 | 1 | 843 |
| QNT66325.1 | 0.0 | 19 | 804 | 31 | 849 |
| ADE83294.1 | 0.0 | 33 | 825 | 33 | 821 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZPS_AAA | 6.79e-72 | 241 | 858 | 2 | 627 | ChainAAA, MgGH51 [Meripilus giganteus],6ZPV_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPW_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPX_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPY_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPZ_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ0_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ1_AAA Chain AAA, MgGH51 [Meripilus giganteus] |
| 3S2C_A | 4.73e-15 | 435 | 763 | 48 | 367 | Structureof the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_B Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_C Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_D Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_E Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_F Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_G Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_H Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_I Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_J Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_K Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_L Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1] |
| 4ATW_A | 6.21e-15 | 435 | 763 | 48 | 367 | Thecrystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_B The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_C The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_D The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_E The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_F The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8] |
| 3UG3_A | 6.77e-15 | 435 | 763 | 68 | 387 | Crystalstructure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG4_A Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG5_A Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P82593 | 4.29e-124 | 248 | 769 | 44 | 556 | Extracellular exo-alpha-L-arabinofuranosidase OS=Streptomyces chartreusis OX=1969 PE=1 SV=1 |
| Q9SG80 | 4.09e-81 | 241 | 760 | 42 | 550 | Alpha-L-arabinofuranosidase 1 OS=Arabidopsis thaliana OX=3702 GN=ASD1 PE=1 SV=1 |
| Q8VZR2 | 4.89e-78 | 241 | 801 | 41 | 589 | Alpha-L-arabinofuranosidase 2 OS=Arabidopsis thaliana OX=3702 GN=ASD2 PE=2 SV=1 |
| Q2U790 | 1.08e-58 | 231 | 850 | 17 | 623 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=abfA PE=3 SV=2 |
| B8NKA3 | 1.48e-58 | 231 | 817 | 17 | 612 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=abfA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
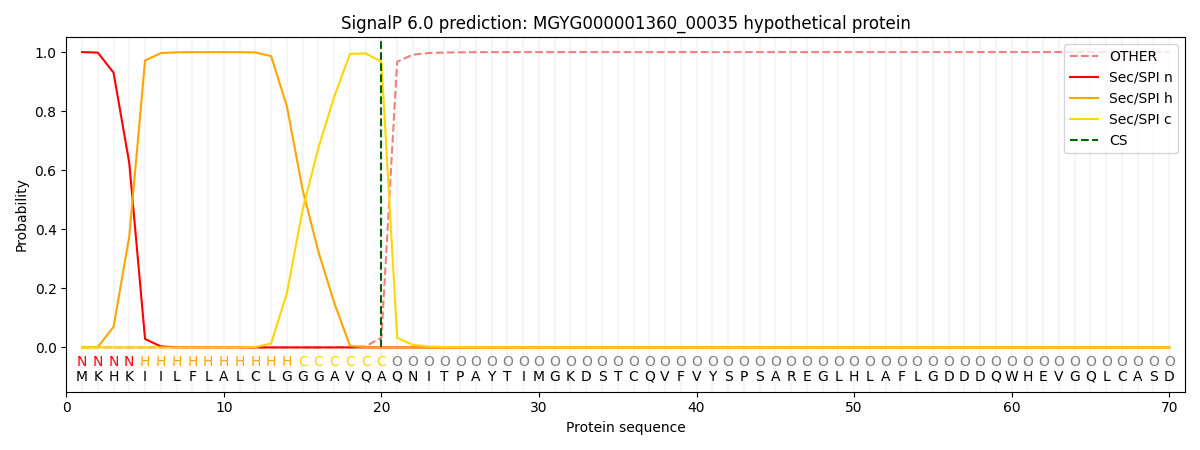
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000317 | 0.999025 | 0.000177 | 0.000166 | 0.000144 | 0.000132 |
