You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002653_00087
You are here: Home > Sequence: MGYG000002653_00087
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp002251295 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp002251295 | |||||||||||
| CAZyme ID | MGYG000002653_00087 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | Intracellular exo-alpha-L-arabinofuranosidase 2 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 103969; End: 105558 Strand: + | |||||||||||
Full Sequence Download help
| MHYHYFLSAA FALCASQVSA ESIKATLKTD QGKNTINREI YGQFAEHLGS CIYGGLWVGE | 60 |
| NSPIPNINGY RKDVFEALKD LNVPVLRWPG GCFADDYHWM DGIGPRDKRP SLRNNNWGGT | 120 |
| IEDNSFGTHE FLNLCEMLGC EPYISGNVGS GTVKEMAQWV EYMTSNGDTP MARLRHQNGR | 180 |
| DKAWHVKYFG IGNEAWGCGG NMTPEYYSDE FRKFNTYLRD QPGNRLYRIG SGASDYDYKW | 240 |
| TSVLMDRIGG RMNGVSLHYY TVTGWNGSKG SAIKFDNDQY YWALGKCLEI EEVIKKHKAI | 300 |
| MDEKDPNHKV DLLVDEWGTW WDEEPGTIAG HLYQQNALRD AFVASLSLDV FHRHTDRVKM | 360 |
| ANIAQVVNVL QSMILTDQEG TGHMVLTPTY YVFKMYAPFQ DASYLPMTIA CDTMKVRHEY | 420 |
| FTPMDAQKAN GYRNLPLLSY SAAKTKDGQI VIALTNVSLD KEQAIDIALE GTNAKSVNGQ | 480 |
| ILTAKNVADF NDFDHPDRVK VADFKQAKLK KNVLSVKIPA KSVVVMTLK | 529 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH51 | 24 | 529 | 7.1e-138 | 0.7253968253968254 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3534 | AbfA | 4.74e-175 | 24 | 529 | 5 | 499 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| pfam06964 | Alpha-L-AF_C | 4.01e-69 | 315 | 522 | 1 | 192 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| smart00813 | Alpha-L-AF_C | 1.23e-59 | 315 | 522 | 1 | 189 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADE81534.1 | 9.55e-317 | 10 | 529 | 11 | 530 |
| QVJ82199.1 | 2.22e-314 | 23 | 529 | 1 | 507 |
| AFN57662.1 | 2.64e-307 | 12 | 529 | 13 | 530 |
| BCS86623.1 | 3.35e-288 | 12 | 529 | 12 | 516 |
| VEH15625.1 | 5.45e-285 | 7 | 529 | 11 | 517 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZT6_A | 2.91e-197 | 24 | 528 | 6 | 493 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT6_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT6_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_A Chain A, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
| 2VRQ_A | 4.13e-197 | 24 | 528 | 6 | 493 | StructureOf An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus],2VRQ_B Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus],2VRQ_C Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus] |
| 6ZT8_A | 1.18e-196 | 24 | 528 | 6 | 493 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT8_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT8_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_A Chain A, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
| 6ZTA_A | 3.87e-195 | 24 | 528 | 6 | 493 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZTA_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZTA_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
| 2VRK_A | 7.29e-188 | 24 | 528 | 6 | 493 | Structureof a seleno-methionyl derivative of wild type arabinofuranosidase from Thermobacillus xylanilyticus [Thermobacillus xylanilyticus],2VRK_B Structure of a seleno-methionyl derivative of wild type arabinofuranosidase from Thermobacillus xylanilyticus [Thermobacillus xylanilyticus],2VRK_C Structure of a seleno-methionyl derivative of wild type arabinofuranosidase from Thermobacillus xylanilyticus [Thermobacillus xylanilyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q59219 | 5.84e-266 | 7 | 528 | 7 | 513 | Intracellular exo-alpha-L-arabinofuranosidase OS=Bacteroides ovatus OX=28116 GN=asdII PE=3 SV=1 |
| P94552 | 6.27e-191 | 24 | 529 | 5 | 492 | Intracellular exo-alpha-L-arabinofuranosidase 2 OS=Bacillus subtilis (strain 168) OX=224308 GN=abf2 PE=1 SV=2 |
| A1CQC3 | 6.92e-68 | 36 | 487 | 26 | 471 | Probable alpha-L-arabinofuranosidase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=abfC PE=3 SV=2 |
| B8NIX4 | 5.07e-67 | 36 | 487 | 26 | 471 | Probable alpha-L-arabinofuranosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=abfC PE=3 SV=1 |
| A2QQ94 | 7.26e-67 | 36 | 487 | 26 | 471 | Probable alpha-L-arabinofuranosidase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=abfC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
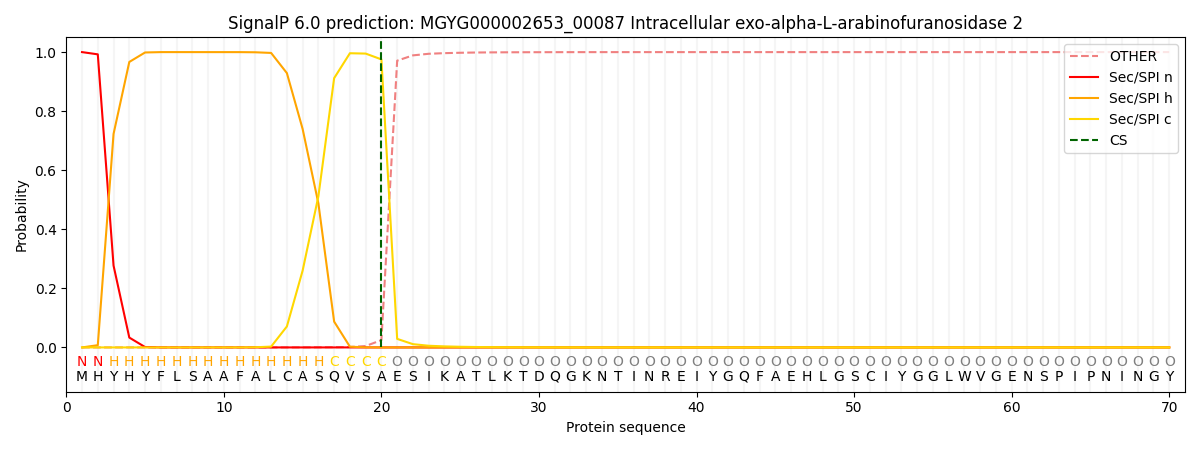
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000273 | 0.999159 | 0.000157 | 0.000148 | 0.000128 | 0.000127 |
