You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001261_00251
You are here: Home > Sequence: MGYG000001261_00251
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Caulobacter sp903900155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Caulobacter; Caulobacter sp903900155 | |||||||||||
| CAZyme ID | MGYG000001261_00251 | |||||||||||
| CAZy Family | GH55 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10594; End: 13605 Strand: - | |||||||||||
Full Sequence Download help
| MRALVLSLLL AASSPVLAAA AAPSSAWPVA PDDPRAVTVK AVGDGKTDDT DAIQKAIDAG | 60 |
| RDSTGHGLVF LPSGRYRLSR TILIPPGIRV FGVGKTRPVL VLGPDTPGFQ DGVGTMVVFT | 120 |
| GGDQYQVGDI PVPVPTVRPA TAKVRDANSG AFYSSMANID IEIGEGNPAA AGVRFRMAQH | 180 |
| AFLKHMNFRL GSGFAGVYQA GNIMEDVHFQ GGRYGIVTEK TSPAWQFTLV DSTFDGQRDA | 240 |
| AIREHEVDLT LVNVAIRNTP VGIEIDKGYS DSLWGKNVRF ENVSKAGVVI SAEDNVFTQV | 300 |
| GFDNAVASNT PVFARFRDSG KTVEGKGKAY RVGSFTYGLT LPAMGQMGEY KTLSDITPLK | 360 |
| ALPAATAPAL RSLPPMAEWT NVKTLGVKGD SSTDDTAALQ KAIDSHKVLY LPIGFYRVSD | 420 |
| TLKLKPDTVL IGLHPALTQL VIPDGNPGHV GVGPVRPILE TPRGGDNIVA GIGLFTGRIN | 480 |
| PRASALLWRS GEKSLVTDVK IMGGGGTPTA DGKPLGAAQA RSGDPVADGR WDAQYPSIWV | 540 |
| TDGGGGAFAN VWSPNTFASA GFYISDTKTP GHIYEVSVEH HVRNEFVLDN VENWEFLAPQ | 600 |
| TEQEVGDGPD AVSLEVRNSR NILFANYHGY RVTRSYHPAE AAVKLFNSTD IRFRNVHINA | 660 |
| ESGIALCDKV GCGTYLRASK YPFENAIQDK TSKLEVRERE FAVLDVKGDQ PAPAPADPRV | 720 |
| KKLETGFWSI SGATTGADGA LYFVEKRFQR IYRWTAAKGL EIVRDASLDP TNLAMDASGK | 780 |
| LLVVSSLGPQ GAVYSIDPGG PHDAMTMIAP TAVTSRGKGK TLLPVNWWNN GEFRDQYDPA | 840 |
| KGEFTTLAQM FARDVGMPKA QEYVSPDGSV SLPAFRVWQQ GPPDHVGWRW SDSLQANGLI | 900 |
| GGAVGERLFF TNGSENKTYS GQVGPGGTLT DLKIFANRGG ESVAVDGQGR VFVANGQIFQ | 960 |
| YAPDGSLTGR IDVPERPLQL IFGGEGGKTL FVLTHHSLYA VTP | 1003 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH55 | 148 | 604 | 3.9e-98 | 0.6418918918918919 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12708 | Pectate_lyase_3 | 2.38e-23 | 41 | 264 | 9 | 213 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| pfam08450 | SGL | 1.52e-12 | 736 | 996 | 9 | 246 | SMP-30/Gluconolaconase/LRE-like region. This family describes a region that is found in proteins expressed by a variety of eukaryotic and prokaryotic species. These proteins include various enzymes, such as senescence marker protein 30 (SMP-30), gluconolactonase and luciferin-regenerating enzyme (LRE). SMP-30 is known to hydrolyze diisopropyl phosphorofluoridate in the liver, and has been noted as having sequence similarity, in the region described in this family, with PON1 and LRE. |
| pfam12708 | Pectate_lyase_3 | 1.08e-11 | 379 | 520 | 1 | 126 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| COG3386 | YvrE | 4.51e-08 | 736 | 997 | 34 | 277 | Sugar lactone lactonase YvrE [Carbohydrate transport and metabolism]. |
| COG5434 | Pgu1 | 0.001 | 40 | 76 | 89 | 124 | Polygalacturonase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADG10334.1 | 0.0 | 1 | 1003 | 1 | 1001 |
| AVQ02066.1 | 0.0 | 1 | 1003 | 1 | 1001 |
| AZS22151.1 | 0.0 | 1 | 1001 | 1 | 1002 |
| AHE56388.1 | 0.0 | 21 | 1003 | 29 | 1014 |
| AUW59624.1 | 0.0 | 21 | 1003 | 24 | 1009 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5M5Z_A | 2.52e-17 | 41 | 651 | 36 | 678 | ChainA, Beta-1,3-glucanase [Thermochaetoides thermophila],5M60_A Chain A, Beta-1,3-glucanase [Thermochaetoides thermophila] |
| 3GQ7_A | 4.46e-06 | 381 | 432 | 24 | 75 | ChainA, Preneck appendage protein [Salasvirus phi29],3GQ8_A Chain A, Preneck appendage protein [Salasvirus phi29],3GQ9_A Chain A, Preneck appendage protein [Salasvirus phi29],3GQA_A Chain A, Preneck appendage protein [Salasvirus phi29] |
| 3SUC_A | 4.93e-06 | 381 | 432 | 24 | 75 | ChainA, Preneck appendage protein [Bacillus phage phi29] |
| 6QZ9_A | 5.10e-06 | 381 | 432 | 106 | 157 | ChainA, Pre-neck appendage protein [Salasvirus phi29],6QZ9_B Chain B, Pre-neck appendage protein [Salasvirus phi29],6QZ9_C Chain C, Pre-neck appendage protein [Salasvirus phi29],6QZ9_D Chain D, Pre-neck appendage protein [Salasvirus phi29],6QZ9_E Chain E, Pre-neck appendage protein [Salasvirus phi29],6QZ9_F Chain F, Pre-neck appendage protein [Salasvirus phi29],6QZ9_G Chain G, Pre-neck appendage protein [Salasvirus phi29],6QZ9_H Chain H, Pre-neck appendage protein [Salasvirus phi29],6QZ9_I Chain I, Pre-neck appendage protein [Salasvirus phi29],6QZ9_J Chain J, Pre-neck appendage protein [Salasvirus phi29],6QZ9_K Chain K, Pre-neck appendage protein [Salasvirus phi29],6QZ9_L Chain L, Pre-neck appendage protein [Salasvirus phi29],6QZ9_M Chain M, Pre-neck appendage protein [Salasvirus phi29],6QZ9_N Chain N, Pre-neck appendage protein [Salasvirus phi29],6QZ9_O Chain O, Pre-neck appendage protein [Salasvirus phi29],6QZ9_P Chain P, Pre-neck appendage protein [Salasvirus phi29],6QZ9_Q Chain Q, Pre-neck appendage protein [Salasvirus phi29],6QZ9_R Chain R, Pre-neck appendage protein [Salasvirus phi29],6QZ9_S Chain S, Pre-neck appendage protein [Salasvirus phi29],6QZ9_T Chain T, Pre-neck appendage protein [Salasvirus phi29],6QZ9_U Chain U, Pre-neck appendage protein [Salasvirus phi29],6QZ9_V Chain V, Pre-neck appendage protein [Salasvirus phi29],6QZ9_W Chain W, Pre-neck appendage protein [Salasvirus phi29],6QZ9_X Chain X, Pre-neck appendage protein [Salasvirus phi29],6QZ9_Y Chain Y, Pre-neck appendage protein [Salasvirus phi29],6QZ9_Z Chain Z, Pre-neck appendage protein [Salasvirus phi29],6QZ9_a Chain a, Pre-neck appendage protein [Salasvirus phi29],6QZ9_b Chain b, Pre-neck appendage protein [Salasvirus phi29],6QZ9_c Chain c, Pre-neck appendage protein [Salasvirus phi29],6QZ9_d Chain d, Pre-neck appendage protein [Salasvirus phi29],6QZ9_e Chain e, Pre-neck appendage protein [Salasvirus phi29],6QZ9_f Chain f, Pre-neck appendage protein [Salasvirus phi29],6QZ9_g Chain g, Pre-neck appendage protein [Salasvirus phi29],6QZ9_h Chain h, Pre-neck appendage protein [Salasvirus phi29],6QZ9_i Chain i, Pre-neck appendage protein [Salasvirus phi29],6QZ9_j Chain j, Pre-neck appendage protein [Salasvirus phi29],6QZF_A Chain A, Pre-neck appendage protein [Salasvirus phi29],6QZF_B Chain B, Pre-neck appendage protein [Salasvirus phi29],6QZF_C Chain C, Pre-neck appendage protein [Salasvirus phi29],6QZF_D Chain D, Pre-neck appendage protein [Salasvirus phi29],6QZF_E Chain E, Pre-neck appendage protein [Salasvirus phi29],6QZF_F Chain F, Pre-neck appendage protein [Salasvirus phi29],6QZF_G Chain G, Pre-neck appendage protein [Salasvirus phi29],6QZF_H Chain H, Pre-neck appendage protein [Salasvirus phi29],6QZF_I Chain I, Pre-neck appendage protein [Salasvirus phi29],6QZF_J Chain J, Pre-neck appendage protein [Salasvirus phi29],6QZF_K Chain K, Pre-neck appendage protein [Salasvirus phi29],6QZF_L Chain L, Pre-neck appendage protein [Salasvirus phi29],6QZF_M Chain M, Pre-neck appendage protein [Salasvirus phi29],6QZF_N Chain N, Pre-neck appendage protein [Salasvirus phi29],6QZF_O Chain O, Pre-neck appendage protein [Salasvirus phi29],6QZF_P Chain P, Pre-neck appendage protein [Salasvirus phi29],6QZF_Q Chain Q, Pre-neck appendage protein [Salasvirus phi29],6QZF_R Chain R, Pre-neck appendage protein [Salasvirus phi29],6QZF_S Chain S, Pre-neck appendage protein [Salasvirus phi29],6QZF_T Chain T, Pre-neck appendage protein [Salasvirus phi29],6QZF_U Chain U, Pre-neck appendage protein [Salasvirus phi29],6QZF_V Chain V, Pre-neck appendage protein [Salasvirus phi29],6QZF_W Chain W, Pre-neck appendage protein [Salasvirus phi29],6QZF_X Chain X, Pre-neck appendage protein [Salasvirus phi29],6QZF_Y Chain Y, Pre-neck appendage protein [Salasvirus phi29],6QZF_Z Chain Z, Pre-neck appendage protein [Salasvirus phi29],6QZF_a Chain a, Pre-neck appendage protein [Salasvirus phi29],6QZF_b Chain b, Pre-neck appendage protein [Salasvirus phi29],6QZF_c Chain c, Pre-neck appendage protein [Salasvirus phi29],6QZF_d Chain d, Pre-neck appendage protein [Salasvirus phi29],6QZF_e Chain e, Pre-neck appendage protein [Salasvirus phi29],6QZF_f Chain f, Pre-neck appendage protein [Salasvirus phi29],6QZF_g Chain g, Pre-neck appendage protein [Salasvirus phi29],6QZF_h Chain h, Pre-neck appendage protein [Salasvirus phi29],6QZF_i Chain i, Pre-neck appendage protein [Salasvirus phi29],6QZF_j Chain j, Pre-neck appendage protein [Salasvirus phi29] |
SignalP and Lipop Annotations help
This protein is predicted as SP
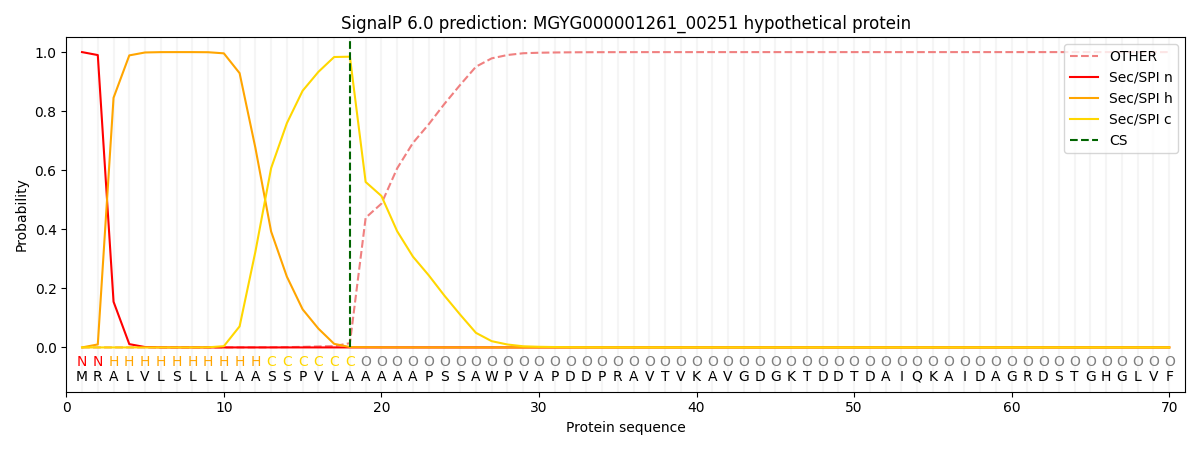
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000454 | 0.998759 | 0.000185 | 0.000220 | 0.000189 | 0.000175 |
