You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001441_00025
You are here: Home > Sequence: MGYG000001441_00025
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pauljensenia sp000411415 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Pauljensenia; Pauljensenia sp000411415 | |||||||||||
| CAZyme ID | MGYG000001441_00025 | |||||||||||
| CAZy Family | GH66 | |||||||||||
| CAZyme Description | Cycloisomaltooligosaccharide glucanotransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 34504; End: 37320 Strand: + | |||||||||||
Full Sequence Download help
| METRHPLTIP AAIVLGTAVV ATALPLPNST PATATGTAHV TRAYTDKSTH EPGKQATITA | 60 |
| EASTEGTVHF SVSHLGTEID SGDATVENGK ATWTYTTPSE DGQGYLVTAT GGDGTHAETA | 120 |
| IDTSTSWTRF PRMGYLSHFK PTAPEGTDGH TTYEGFLFQK PQDYIDKLSR DYHINALQYY | 180 |
| DWQYRHEQPV ATGDLADKWP LWYRDTYASK KTINDYIQDA GKANIGSLAY SMAYAANDNY | 240 |
| DTNTIKDEWR LREDNGSYWV RDLGEQWWVP TPKGVDKPKS HQFMMNVNNE DWRTYITGQY | 300 |
| KTQKTEFKFD GTHIDTLGQT SKKDASGNPV DLTDGLAALV DDTYKNVGGQ VGINLPDGAG | 360 |
| SEKINKASAA YMYTELWDHN ETNAQVASYL QSARNNAGNK PQIVAAYANN YNPTISLPDP | 420 |
| KDSKKTIHPG VTPDEGMRIE AESDQASVSG GAQILSGDDS ASGGAYAGNF SAGGSTVTFT | 480 |
| VDAGQGGTFT FTTRYARQDA DGAYHQMILD MGKSGQQKLI KYVHFDQTGS YYTWKDMTET | 540 |
| VELTPGIHTI SYWVPNDKNY TPVNIDCITF REFNTDSVKL ADAAFAANGA HHLELGDYGR | 600 |
| MLDNEFFVNS GRSMSTDLQT WMKNYYNIST AYENLLFGDN LTRKERQVEV STNGVGLPTS | 660 |
| TDGSANTIWA NTMTSNAGTA LHLINLRTND SEGNDEYWRN AAKQILAFDN TSVTYHLEDG | 720 |
| EAIPGSIFAV SPDVDGGRPM PLDFTTGTDE QGRTTLTFNV GRLSSWDMVV FSPTKVADRA | 780 |
| DAAPQDPNAS GNANSDNAGL VPATVVGQLR NGHGQCLTSQ DPKGADGTPV WNSNCAGNSA | 840 |
| AQIVTYEGDG HLRIGDRCVD VVGGYTNEGT VAHMWTCYPT LESQKWDING SGQLVNRASG | 900 |
| LCLTIPGDTT QENTQAVISQ CSDASTSQRW TLTNVSGR | 938 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH66 | 42 | 411 | 7.7e-69 | 0.6420863309352518 |
| CBM13 | 810 | 935 | 4.4e-20 | 0.6702127659574468 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13199 | Glyco_hydro_66 | 1.98e-94 | 45 | 770 | 1 | 557 | Glycosyl hydrolase family 66. This family is a set of glycosyl hydrolase enzymes including cycloisomaltooligosaccharide glucanotransferase (EC:2.4.1.-) and dextranase (EC:3.2.1.11) activities. |
| cd14745 | GH66 | 1.15e-85 | 129 | 632 | 1 | 331 | Glycoside Hydrolase Family 66. Glycoside Hydrolase Family 66 contains proteins characterized as cycloisomaltooligosaccharide glucanotransferase (CITase) and dextranases from a variety of bacteria. CITase cyclizes part of a (1-6)-alpha-D-glucan (dextrans) chain by formation of a (1-6)-alpha-D-glucosidic bond. Dextranases catalyze the endohydrolysis of (1-6)-alpha-D-glucosidic linkages in dextran. Some members contain Carbohydrate Binding Module 35 (CBM35) domains, either C-terminal or inserted in the domain or both. |
| pfam00652 | Ricin_B_lectin | 2.74e-24 | 806 | 930 | 2 | 126 | Ricin-type beta-trefoil lectin domain. |
| cd00161 | RICIN | 1.47e-21 | 808 | 932 | 4 | 124 | Ricin-type beta-trefoil; Carbohydrate-binding domain formed from presumed gene triplication. The domain is found in a variety of molecules serving diverse functions such as enzymatic activity, inhibitory toxicity and signal transduction. Highly specific ligand binding occurs on exposed surfaces of the compact domain sturcture. |
| NF035929 | lectin_1 | 3.81e-17 | 755 | 932 | 675 | 836 | lectin. Lectins are important adhesin proteins, which bind carbohydrate structures on host cell surface. The carbohydrate specificity of diverse lectins to a large extent dictates bacteria tissue tropism by mediating specific attachment to unique host sites expressing the corresponding carbohydrate receptor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCT35775.1 | 0.0 | 1 | 901 | 1 | 903 |
| QIS74997.1 | 4.76e-114 | 4 | 837 | 5 | 648 |
| QGS11685.1 | 2.23e-104 | 38 | 847 | 34 | 767 |
| QNK58819.1 | 4.23e-89 | 32 | 769 | 32 | 732 |
| BAA09604.1 | 4.26e-88 | 32 | 774 | 36 | 741 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3WNK_A | 6.16e-91 | 36 | 769 | 23 | 719 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 3WNP_A | 9.23e-91 | 36 | 769 | 4 | 700 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNP_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 3WNL_A | 6.58e-90 | 36 | 769 | 4 | 700 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNM_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNN_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNN_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNO_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNO_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 5X7G_A | 1.50e-86 | 40 | 769 | 26 | 718 | CrystalStructure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase [Paenibacillus sp. 598K],5X7H_A Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltoheptaose [Paenibacillus sp. 598K] |
| 5AXG_A | 1.39e-44 | 44 | 769 | 48 | 606 | Crystalstructure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus [Thermoanaerobacter pseudethanolicus ATCC 33223],5AXG_B Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus [Thermoanaerobacter pseudethanolicus ATCC 33223] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94286 | 8.51e-89 | 32 | 774 | 36 | 741 | Cycloisomaltooligosaccharide glucanotransferase OS=Niallia circulans OX=1397 PE=1 SV=1 |
| P70873 | 2.06e-84 | 40 | 786 | 36 | 739 | Cycloisomaltooligosaccharide glucanotransferase OS=Niallia circulans OX=1397 GN=cit PE=3 SV=1 |
| Q59979 | 6.06e-10 | 67 | 769 | 2 | 595 | Dextranase OS=Streptococcus salivarius OX=1304 GN=dex PE=3 SV=1 |
| P39653 | 1.63e-09 | 571 | 772 | 581 | 801 | Dextranase OS=Streptococcus downei OX=1317 GN=dex PE=1 SV=1 |
| Q54443 | 1.63e-08 | 557 | 774 | 500 | 735 | Dextranase OS=Streptococcus mutans serotype c (strain ATCC 700610 / UA159) OX=210007 GN=dexA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
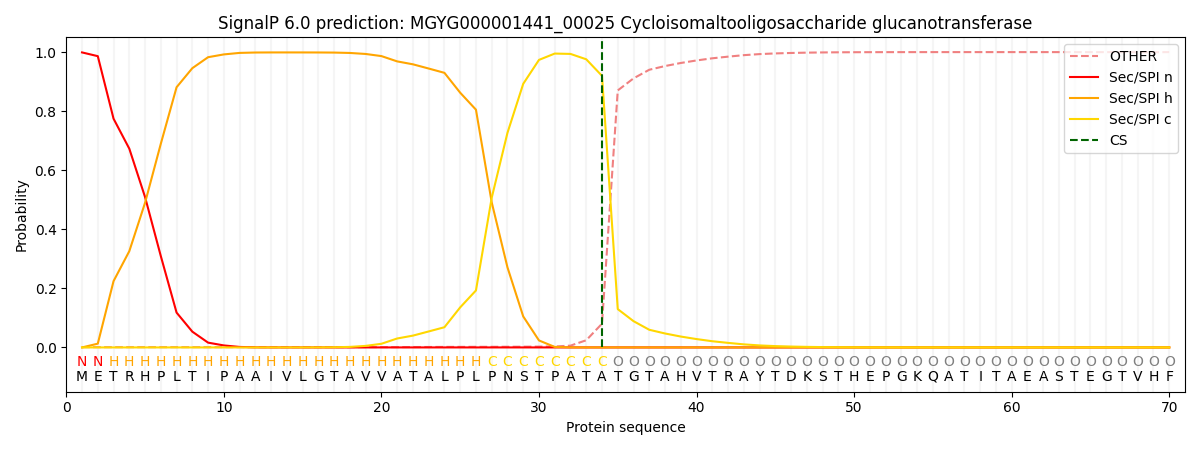
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002140 | 0.996574 | 0.000375 | 0.000398 | 0.000259 | 0.000219 |
