You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000010_02539
You are here: Home > Sequence: MGYG000000010_02539
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Staphylococcus_A lentus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus_A; Staphylococcus_A lentus | |||||||||||
| CAZyme ID | MGYG000000010_02539 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10778; End: 14767 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 1175 | 1308 | 4.5e-20 | 0.9453125 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 9.78e-70 | 1070 | 1328 | 1 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| COG5632 | CwlA | 4.12e-31 | 462 | 746 | 1 | 279 | N-acetylmuramoyl-L-alanine amidase CwlA [Cell wall/membrane/envelope biogenesis]. |
| smart00047 | LYZ2 | 1.32e-30 | 1164 | 1323 | 2 | 147 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| COG5271 | MDN1 | 6.87e-25 | 99 | 420 | 3792 | 4123 | Midasin, AAA ATPase with vWA domain, involved in ribosome maturation [Translation, ribosomal structure and biogenesis]. |
| smart00644 | Ami_2 | 2.22e-23 | 480 | 608 | 1 | 126 | Ami_2 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMU10569.1 | 0.0 | 1 | 1329 | 1 | 1392 |
| SQE51070.1 | 0.0 | 1 | 1329 | 1 | 1313 |
| QDR64573.1 | 0.0 | 401 | 1329 | 405 | 1317 |
| QSN66810.1 | 0.0 | 1 | 1329 | 1 | 1296 |
| QRN90500.1 | 0.0 | 406 | 1329 | 478 | 1384 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4KNK_A | 6.08e-79 | 439 | 635 | 20 | 216 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNK_B Chain B, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_A Chain A, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_B Chain B, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_C Chain C, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_D Chain D, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 3LAT_A | 4.04e-76 | 439 | 634 | 11 | 206 | Crystalstructure of Staphylococcus peptidoglycan hydrolase AmiE [Staphylococcus epidermidis],3LAT_B Crystal structure of Staphylococcus peptidoglycan hydrolase AmiE [Staphylococcus epidermidis] |
| 6FXO_A | 2.12e-75 | 1095 | 1328 | 13 | 244 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXP_A | 1.15e-54 | 1076 | 1328 | 2 | 246 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50],6FXP_B Chain B, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50] |
| 6U0O_B | 2.24e-54 | 1095 | 1328 | 51 | 276 | ChainB, LYZ2 domain-containing protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6GAG0 | 2.62e-158 | 439 | 1328 | 216 | 1250 | Bifunctional autolysin OS=Staphylococcus aureus (strain MSSA476) OX=282459 GN=atl PE=3 SV=1 |
| Q99V41 | 3.53e-157 | 399 | 1328 | 159 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
| Q931U5 | 3.53e-157 | 399 | 1328 | 159 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=atl PE=1 SV=2 |
| A7X0T9 | 4.12e-157 | 399 | 1328 | 166 | 1255 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu3 / ATCC 700698) OX=418127 GN=atl PE=3 SV=2 |
| P0C5Z8 | 4.12e-157 | 399 | 1328 | 166 | 1255 | Bifunctional autolysin OS=Staphylococcus aureus OX=1280 GN=atl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
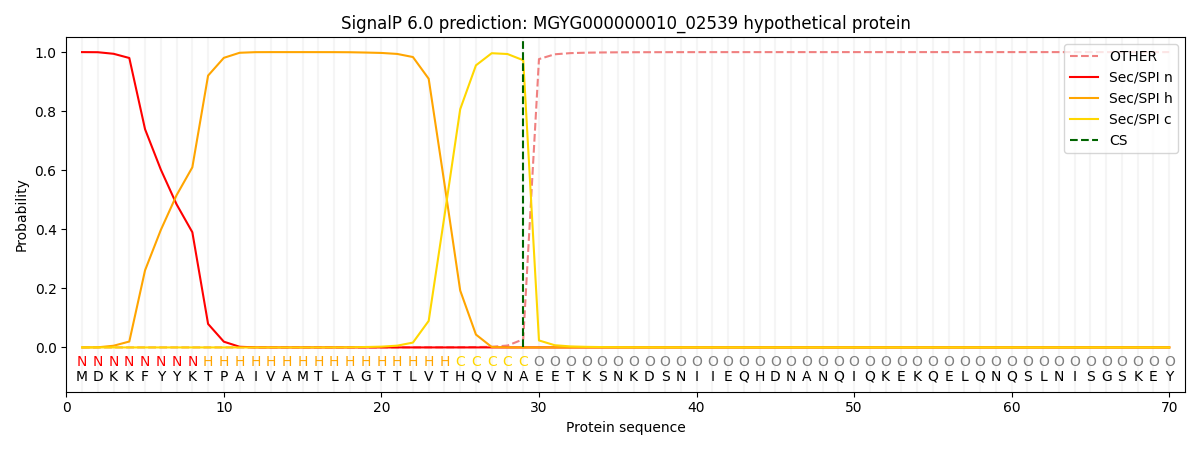
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000379 | 0.998858 | 0.000263 | 0.000166 | 0.000157 | 0.000143 |
