You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001008_00478
You are here: Home > Sequence: MGYG000001008_00478
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium_AQ sp900552125 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; Clostridium_AQ; Clostridium_AQ sp900552125 | |||||||||||
| CAZyme ID | MGYG000001008_00478 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24213; End: 26003 Strand: + | |||||||||||
Full Sequence Download help
| MKKTITFIAV FLCGYLLLLQ QNNWLSAQIT NEKQPEQVGA TLSGDVILKH IDENGKIIID | 60 |
| DTEELEAITK QEQAETERLS KIKGIGGAKN ISYGVVNFKT KSSANYNTRY TDVATGQSGY | 120 |
| TNGYYGADGA FLGYTNNGTK VKFMLSGVVG LVDADEVEVL DYEDESTVKS VNYYYCKNGN | 180 |
| IYHYITLNVR QPYYSSVLMV GKQQSYMKSN TVYYSYDGHY FYTTYEKMIN DYKADTRKNS | 240 |
| INPSNPYYNY YQYVSTRTKT NFTAADFNGY VKSYLGSSYN ASNTKMYEMG KYFIDYQNTY | 300 |
| GAYALSTFGV AVNESYFGTS SIALSKNNLF GHNAVDSDPG LANGYSSPQN SIRDHAKYYV | 360 |
| NLWYSTPKYS SYHGSYLGDK AGGMNVSYAS DPYWGETAAH WMWQLDEYSK NSDANSKTLV | 420 |
| VKDNSSVNIR KEATTSSSSL YTTSENIMSF ILLDTVKGDA VSGNTTWYKV QLDTPLNSSR | 480 |
| TGIDYSGNSY NFSKSYGYIH SSLLTKVNSG TDSSGGNSGG SSGGNTGSSA YKLGDVNDDG | 540 |
| KISPADYVKV KNHIMGRAIL KDNSLKAADV NKDGKISPAD YVKIKNYIMG KDTIKE | 596 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 289 | 405 | 7.3e-17 | 0.9765625 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14256 | Dockerin_I | 4.23e-16 | 533 | 588 | 1 | 56 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam00404 | Dockerin_1 | 1.78e-13 | 534 | 589 | 1 | 56 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| pfam01832 | Glucosaminidase | 3.83e-08 | 290 | 355 | 1 | 77 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| cd14252 | Dockerin_like | 2.97e-07 | 534 | 588 | 1 | 56 | Dockerin repeat domains and domains resembling dockerin repeats. Dockerins are modules in the cellulosome complex that often anchor catalytic subunits by binding to cohesin domains of scaffolding proteins. Three types of dockerins and their corresponding cohesin have been described in the literature. This alignment models two consecutive dockerin repeats, the functional unit. |
| COG4193 | LytD | 1.73e-06 | 231 | 407 | 50 | 230 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJA02388.1 | 7.02e-291 | 2 | 565 | 3 | 561 |
| QSI26463.1 | 2.24e-284 | 2 | 565 | 3 | 559 |
| QIX08957.1 | 4.11e-278 | 2 | 565 | 3 | 561 |
| ANU71450.1 | 8.27e-278 | 2 | 565 | 3 | 561 |
| ASU21015.1 | 8.27e-278 | 2 | 565 | 3 | 561 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 1.60e-12 | 125 | 407 | 31 | 284 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 1.91e-13 | 125 | 407 | 399 | 652 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 2.01e-13 | 125 | 407 | 443 | 696 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
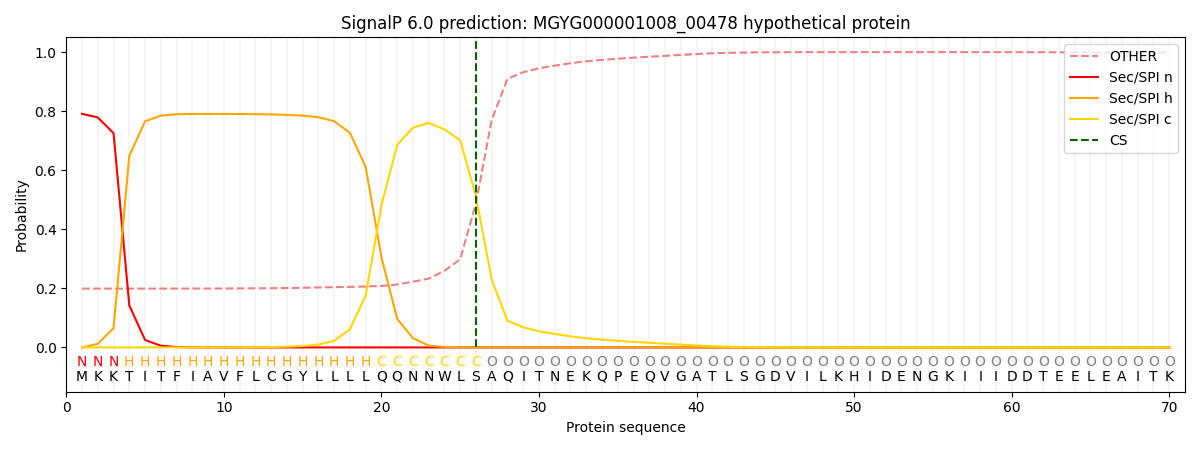
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.220458 | 0.765863 | 0.012092 | 0.000518 | 0.000416 | 0.000621 |
