You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002234_00116
You are here: Home > Sequence: MGYG000002234_00116
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bariatricus sp900554415 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Bariatricus; Bariatricus sp900554415 | |||||||||||
| CAZyme ID | MGYG000002234_00116 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 102649; End: 103419 Strand: + | |||||||||||
Full Sequence Download help
| MRKILLSVNL LMFLACVYVV SPKAANYIDI PWYGKMEESG VQQEQTDDTQ TDEGNTEDGT | 60 |
| ANVTDNEAED AASVEVIVEG QYPIMGQTDI TAEQMAAYFR QSGAAYPAEV LSAGGAPDIE | 120 |
| TFCQIYCEEA EKEGVRSEVA FAQTMKETGW LQYGGDAEVT QYNFAGLGTT GNGVQGNSYS | 180 |
| DVREGVRAQI QHLKAYATEE ALNSDCVDNR YSYVTKGSAP YVEWLGQQEN PEGYGWATGE | 240 |
| NYGYDIVKMI QEMKNM | 256 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 123 | 254 | 2.4e-21 | 0.9765625 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01832 | Glucosaminidase | 1.00e-08 | 123 | 189 | 1 | 77 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRT51288.1 | 1.93e-94 | 73 | 254 | 71 | 253 |
| QHB24702.1 | 4.76e-92 | 69 | 254 | 73 | 258 |
| QEI32209.1 | 4.76e-92 | 69 | 254 | 73 | 258 |
| QRT30849.1 | 4.76e-92 | 69 | 254 | 73 | 258 |
| CBL25595.1 | 7.79e-89 | 36 | 253 | 37 | 258 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O51481 | 2.33e-23 | 119 | 253 | 68 | 195 | Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
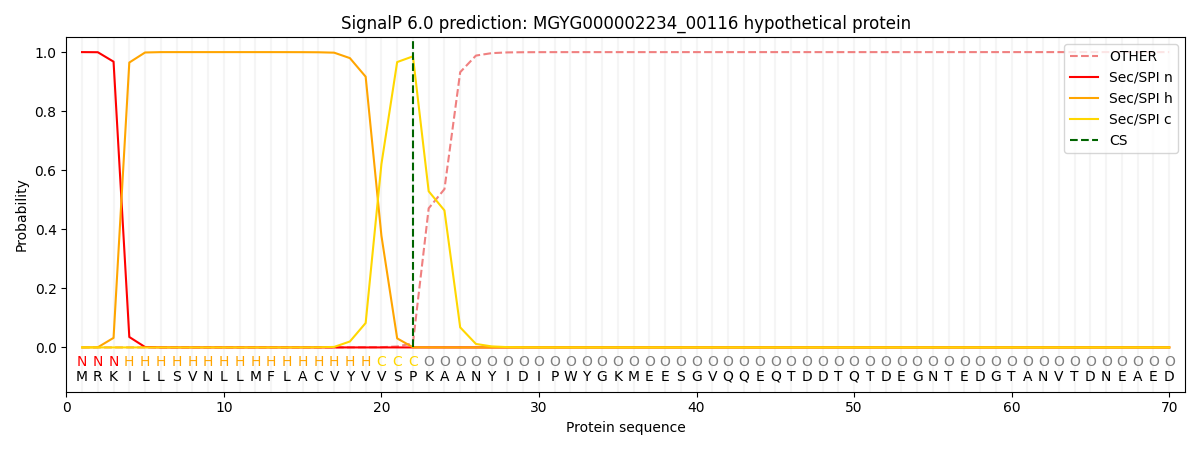
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000219 | 0.999220 | 0.000159 | 0.000134 | 0.000123 | 0.000122 |
