You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002400_00214
You are here: Home > Sequence: MGYG000002400_00214
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Staphylococcus lugdunensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus; Staphylococcus lugdunensis | |||||||||||
| CAZyme ID | MGYG000002400_00214 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | N-acetylmuramoyl-L-alanine amidase domain-containing protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 31457; End: 33463 Strand: - | |||||||||||
Full Sequence Download help
| MSKQKIFIYL LSATLIVPTF TSDIAHATEL TNQSHNESNH ASVDKSDNDD TSDTNDEQTT | 60 |
| KKQEKSAKTT KFKQYTRAKK ASHDEDDLSI FNKLFSDDTL SSNYLNPRGQ QDSLSELLTA | 120 |
| IFSSDSETTE RTAQDTSDDM EPAAEQQDKA STTDSTEQHS TTTSEQNDQS AKDATTDDNK | 180 |
| VSDALDELDN LDDASNHAQD TQRRHDSEAS KDDDKETSSE EKPHQEAPSH DTAQDKQNDS | 240 |
| NKESTDSASD AVIDSILDEY SDNAKKTQQD YTQDKTSDKH KQSNATTTTH QKYAHIDVKK | 300 |
| DRETRHQIDN PQLPSKDSVS HATVPAQSFE EATKQANTRA TSLFATIPQA SDNTSSTDDL | 360 |
| SVVDSQQTRT FIKAIGTDAH QLGQENDIYA SVMIAQAILE SDSGQSTLAK APNFNLFGIK | 420 |
| GAYKGESSEF NTLEATANNQ LYSIQAAFRK YPNKKASLED YVQLIKEGID GNATIYQPTW | 480 |
| KSETSSYRDA TAHLARTYAT DPNYATKLNQ LIKHYHLTDF DHKKLPDLTH YNSSNPGMDN | 540 |
| TGNDFKPFID TNSTSPYPQG QCTWYVYKRM QQFDLAIGGD LGDAHNWNNR AQSEGYHVDT | 600 |
| TPKTHSAVVF EAGQAGASSY YGHVAFVERV NDDGSIVISE SNVKGLGIIS YRTLDANTAD | 660 |
| QLSYISGK | 668 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 377 | 517 | 9.9e-28 | 0.9765625 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK08581 | PRK08581 | 0.0 | 1 | 668 | 1 | 619 | amidase domain-containing protein. |
| COG1705 | FlgJ | 3.38e-55 | 353 | 521 | 28 | 188 | Flagellum-specific peptidoglycan hydrolase FlgJ [Cell wall/membrane/envelope biogenesis, Cell motility]. |
| PRK06347 | PRK06347 | 5.14e-45 | 281 | 543 | 63 | 323 | 1,4-beta-N-acetylmuramoylhydrolase. |
| COG3942 | COG3942 | 2.96e-36 | 526 | 668 | 37 | 173 | Surface antigen [Cell wall/membrane/envelope biogenesis]. |
| smart00047 | LYZ2 | 2.75e-28 | 365 | 515 | 5 | 139 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARJ10433.1 | 0.0 | 1 | 668 | 1 | 668 |
| ATG70168.1 | 0.0 | 1 | 668 | 1 | 668 |
| QRF16767.1 | 0.0 | 1 | 668 | 1 | 668 |
| AMG61918.1 | 0.0 | 1 | 668 | 1 | 668 |
| ATN15415.1 | 0.0 | 1 | 668 | 1 | 668 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5T1Q_A | 1.04e-161 | 310 | 668 | 3 | 359 | ChainA, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_B Chain B, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_C Chain C, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_D Chain D, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 3FI7_A | 1.05e-38 | 369 | 521 | 31 | 183 | CrystalStructure of the autolysin Auto (Lmo1076) from Listeria monocytogenes, catalytic domain [Listeria monocytogenes EGD-e] |
| 2K3A_A | 1.07e-18 | 553 | 665 | 48 | 152 | ChainA, CHAP domain protein [Staphylococcus saprophyticus subsp. saprophyticus ATCC 15305 = NCTC 7292] |
| 2LRJ_A | 2.67e-17 | 551 | 665 | 5 | 111 | ChainA, Staphyloxanthin biosynthesis protein, putative [Staphylococcus aureus subsp. aureus COL] |
| 5DN5_A | 2.06e-11 | 368 | 513 | 3 | 146 | Structureof a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],5DN5_B Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],5DN5_C Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q2G222 | 3.17e-161 | 299 | 668 | 252 | 619 | N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=SAOUHSC_02979 PE=1 SV=1 |
| P39046 | 1.65e-33 | 368 | 521 | 62 | 214 | Muramidase-2 OS=Enterococcus hirae (strain ATCC 9790 / DSM 20160 / JCM 8729 / LMG 6399 / NBRC 3181 / NCIMB 6459 / NCDO 1258 / NCTC 12367 / WDCM 00089 / R) OX=768486 GN=EHR_05900 PE=1 SV=1 |
| P37710 | 3.10e-33 | 365 | 588 | 177 | 389 | Autolysin OS=Enterococcus faecalis (strain ATCC 700802 / V583) OX=226185 GN=EF_0799 PE=1 SV=2 |
| Q9CIT4 | 4.41e-30 | 365 | 521 | 59 | 214 | Probable N-acetylmuramidase OS=Lactococcus lactis subsp. lactis (strain IL1403) OX=272623 GN=acmA PE=3 SV=1 |
| P0C2T5 | 7.82e-30 | 365 | 521 | 59 | 214 | Probable N-acetylmuramidase OS=Lactococcus lactis subsp. cremoris OX=1359 GN=acmA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
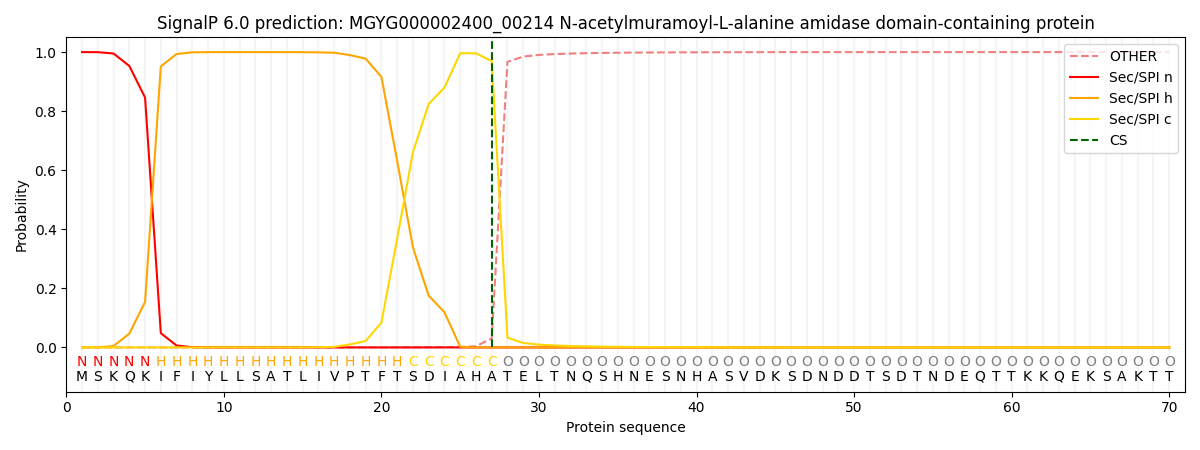
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000328 | 0.998956 | 0.000175 | 0.000176 | 0.000163 | 0.000141 |
