You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002445_00506
You are here: Home > Sequence: MGYG000002445_00506
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
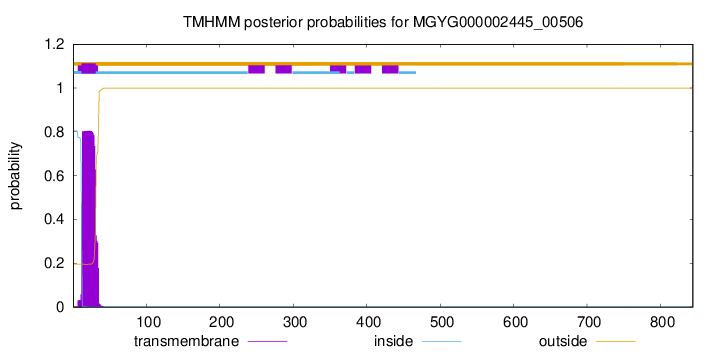
TMHMM annotations
Basic Information help
| Species | Clostridium_Q sp003024715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Clostridium_Q; Clostridium_Q sp003024715 | |||||||||||
| CAZyme ID | MGYG000002445_00506 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 558012; End: 560546 Strand: - | |||||||||||
Full Sequence Download help
| MKKWKLLGKR AAAFLLGAVM AVNPGLAGTA AESLLGIVSY AEERTASVNA TNLNVRSGPG | 60 |
| TSYQALAKLS KGAPVTVLGE QTGTDGVVWY QIRYSGSGGE QTGYVSSKYI KFPTAIKSDS | 120 |
| DFESYLTKQG FPESYKQGLR ELHAQYPNWT FTAFQTNLDW NTALKNESVI GRNLVASSSI | 180 |
| SSWKSTETGA YDWGTGTWPG FDGSSWVQAS SDIIAYYMDP RNFLNDKYIY QFMKQSYDSS | 240 |
| LHTKTGLQNM VSGTFLSGTV SGGSSVSAGT GSSSGSSSSG SATGPGSSSG SSGSSAGLSS | 300 |
| PIASISDHKT ERVAATNGPG MAGDGFSSNN ADGETAPVSS GTSNKSSGTT SSTAGTTSGS | 360 |
| TSAPGSSTTG TTSGSTSSGR TSAPGSGTYS SSGTSTGVAQ QSAIGTSYVD IIMNAASESG | 420 |
| VNPYILASMI LQEQGSQGTG RSISGTVSGY QGYYNFFNIE AYQSGSMSAV ERGLWWASQS | 480 |
| GSYLRPWNSI EKSIRGGSVY YGENYVKKGQ DTLYLKKFNV QGSNLYKHQY MTNVQAAAAE | 540 |
| GLTLSKITAL ANTTLEFSIP VYKNMPAGAC VKPTLDGSPN NKLSALGVDG FALTPTFNRD | 600 |
| TESYDLIVDP SVASVTVAAS AIDSKATIAG TGTVDLQSGV NEITVTVKAA NGNLRNYVLH | 660 |
| VVRQSGGPTY SSSIGSGVSS GSNSSGPATG PGMSSGSSSG NSSSGGSDVQ IVGPTSSGSS | 720 |
| SGSSPSGQSA PGSNVNGFGS GSSGTLIAPN GESVGSAGSP QGSQSPTSAA APSQSTSGTH | 780 |
| AASSVRGDVN GDGKLNIMDI LLAQRSILGL GKLRAEEIAR ADLNGNGKVD ILDVLLMQKK | 840 |
| ILGL | 844 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14256 | Dockerin_I | 1.37e-18 | 786 | 842 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam00404 | Dockerin_1 | 2.17e-13 | 787 | 842 | 1 | 56 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| COG4193 | LytD | 7.00e-13 | 415 | 563 | 106 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam08239 | SH3_3 | 4.55e-12 | 51 | 111 | 1 | 54 | Bacterial SH3 domain. |
| smart00287 | SH3b | 1.20e-10 | 43 | 108 | 1 | 59 | Bacterial SH3 domain homologues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK77705.1 | 1.18e-251 | 9 | 813 | 7 | 837 |
| QRV21758.1 | 6.67e-213 | 32 | 810 | 21 | 746 |
| ADL03932.1 | 6.67e-213 | 32 | 810 | 21 | 746 |
| ANU49989.1 | 5.74e-208 | 9 | 673 | 7 | 687 |
| QQR01104.1 | 5.74e-208 | 9 | 673 | 7 | 687 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FXO_A | 1.05e-08 | 415 | 563 | 98 | 244 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 2KQ8_A | 8.25e-08 | 48 | 110 | 7 | 60 | ChainA, Cell wall hydrolase [[Bacillus thuringiensis] serovar konkukian] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8CPQ1 | 4.15e-09 | 415 | 563 | 1189 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| O33635 | 9.39e-09 | 415 | 563 | 1189 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q5HQB9 | 9.39e-09 | 415 | 563 | 1189 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| Q931U5 | 3.17e-07 | 415 | 563 | 1102 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=atl PE=1 SV=2 |
| Q99V41 | 3.17e-07 | 415 | 563 | 1102 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
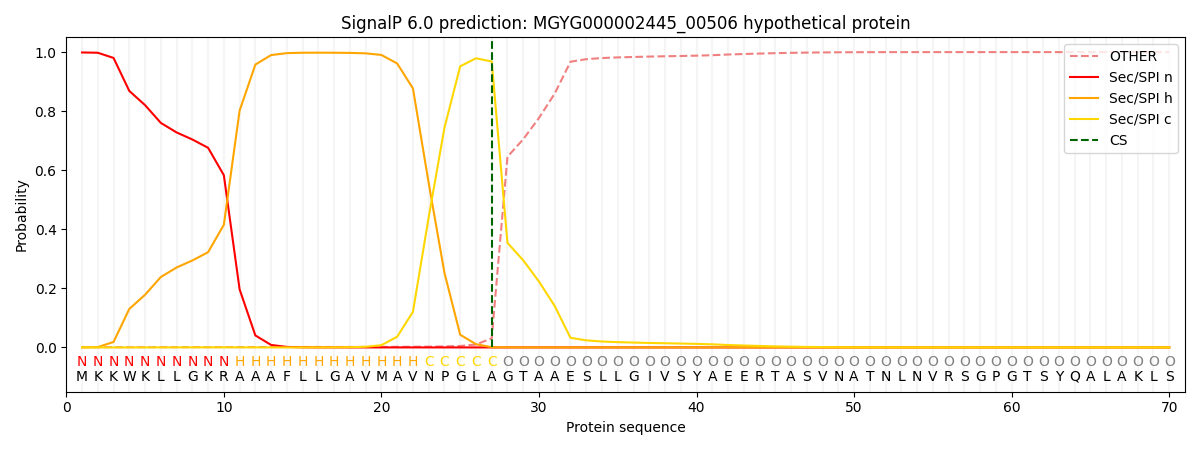
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002027 | 0.996535 | 0.000699 | 0.000259 | 0.000219 | 0.000202 |