You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002945_00358
You are here: Home > Sequence: MGYG000002945_00358
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
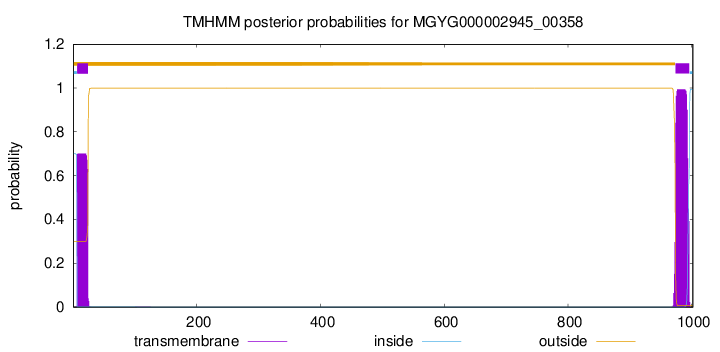
TMHMM annotations
Basic Information help
| Species | Bariatricus massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Bariatricus; Bariatricus massiliensis | |||||||||||
| CAZyme ID | MGYG000002945_00358 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 362996; End: 366004 Strand: - | |||||||||||
Full Sequence Download help
| MSQKRKLLIL SLFLCLSISS VFGTKSKAED GSGEQTSSTS EMKDENAQRL ESVTGMDEDG | 60 |
| NIYEVDDSEG TLESSGIARY LRSVSLKVVN FRTKGNATTN YTEVGTGVSG YTNGAYGADA | 120 |
| AYLGTSGGKV KFMLAGVVGQ VDESEVQVVN FSEAKSVSHY KVSNGRLVHY ITQNMSMTSG | 180 |
| STGSTLDNGP APSYLSSGTT YYSYDGHYFY TNYGTMLSDY QNDSRSQSVN ADSPFYNYFQ | 240 |
| YLPMRSKTGY SGAELNTIVN NRTSAGSKMR DAGNQFVSNQ DIYGVNALLM AGIAANESAW | 300 |
| GNSNICQTKN NLFGLNAVDS SPEASADVYA SVNECIRQFS NGWMSRQYLN PSNWKYKGAF | 360 |
| LGNKASGINV SYASDPYWGE KAAAIAWKLD GDGGSRDNGA YTFAVKDTIS TSHVSVNVRN | 420 |
| ESNTSCSVVY KTGGHSSYSV LVLDSNPVNN FYRIQSDAVL NSGRTGIASG VGEYNFDNMY | 480 |
| AYISADYVTI VNQGNNVPAK KELQSISIST PPSKTAYTEG ETFDAAGMKV LAQWSDGSET | 540 |
| DITQSAAYPS EPLALGTTSI TVSYTFDGVT KTAGQAVQVI EKAVISGVEV NPQSVEMKPG | 600 |
| ESFTFGVLVT GTGDFSKNVT WRVEGAVSAD TKIDGNGKLE IASEESAESL TVKVLSDADE | 660 |
| TKWAQASVTV IKDSTTPEPE NPDDSVAPDP ENPDTEKPDI ENPGTIEAKD ETTGIKVSGT | 720 |
| MPEGVKLEVQ TITQDDEHYE ALVSDERVKS NTILGVFDIK LDKDFDFSQP LSLSFPVDTA | 780 |
| YNGQEVIILH YYEDDGKTFT QSFTASVEEG MATIEVNGFS PYVLALNDAP VPPADEPADE | 840 |
| PADTIPPADT TPPADEPADE PADTVPPADE PADTTPPSDI TPPANTIPPT DEPTDTIPPA | 900 |
| DTTPPAFESN TPPTGENTVT PPTGDGTTAE STDVPPETVI KEQNKQYKGT SGDSQTTTVK | 960 |
| EQSKAPKTGD TSSAVIWVLL LAASAITVLT AGYGKVRRKL HK | 1002 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.84e-15 | 219 | 389 | 50 | 228 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam01832 | Glucosaminidase | 7.03e-11 | 273 | 337 | 1 | 75 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| PHA03291 | PHA03291 | 1.90e-09 | 810 | 935 | 162 | 274 | envelope glycoprotein I; Provisional |
| NF033761 | gliding_GltJ | 5.04e-09 | 828 | 938 | 398 | 516 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| NF033761 | gliding_GltJ | 2.07e-08 | 829 | 953 | 413 | 540 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEK17141.1 | 0.0 | 1 | 836 | 1 | 858 |
| AWY97555.1 | 2.07e-202 | 3 | 672 | 2 | 683 |
| QOV20091.1 | 4.49e-147 | 3 | 495 | 5 | 480 |
| QPS13527.1 | 1.18e-114 | 35 | 541 | 55 | 568 |
| QMW73215.1 | 1.18e-114 | 35 | 541 | 55 | 568 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 3.90e-28 | 129 | 397 | 59 | 290 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 5.68e-26 | 129 | 397 | 427 | 658 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 6.78e-26 | 129 | 397 | 471 | 702 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
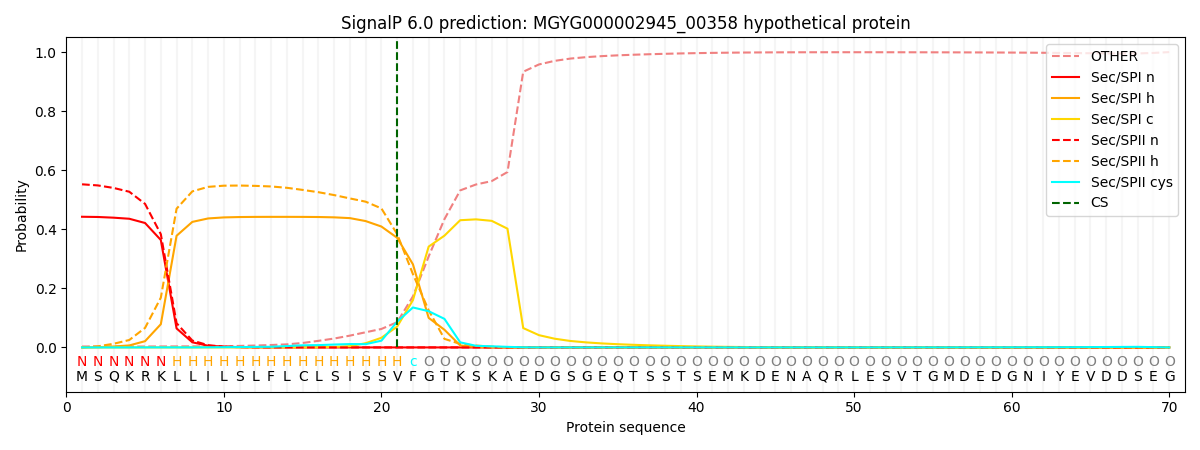
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.005094 | 0.430780 | 0.560322 | 0.002724 | 0.000716 | 0.000331 |