You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002997_00026
You are here: Home > Sequence: MGYG000002997_00026
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
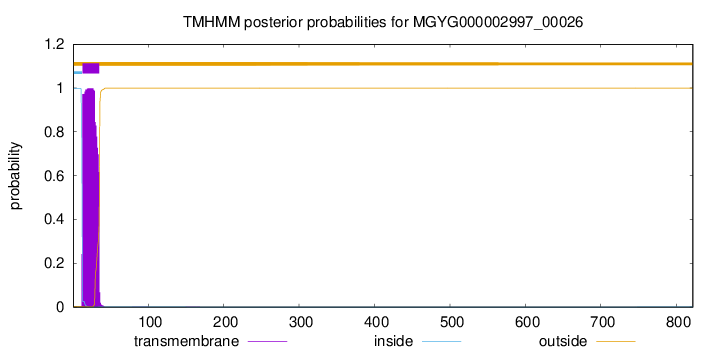
TMHMM annotations
Basic Information help
| Species | Lactococcus_A piscium_C | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Lactococcus_A; Lactococcus_A piscium_C | |||||||||||
| CAZyme ID | MGYG000002997_00026 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 27248; End: 29716 Strand: + | |||||||||||
Full Sequence Download help
| MYYSNCREKQ RNIQVVVAIV LLIGMFISIK VSASAVNDYI INNNIKPANE TLQLGRIYNQ | 60 |
| DPKINGNLNM NYDDGKPKMI IIHEIGVDGG TINGSIDYMV RTQDSAFVHA FVDDSRLITI | 120 |
| ADKSKKSWGS GGYGNRYGIQ IEQMRVTSKA AFYKQIATLA NWTAQQMVKY DMGAPKLMSS | 180 |
| PSTPQRNDLS TKPDGNLATH KMISYKYNQT TDHVDPDEYW ARFGYDINQF RDLVATYYNG | 240 |
| NQVKQNVGYL DTLGITGEGN TIKVRGWHYS PKKYEFLFIM DADTGKEISR QQVSQPEIRN | 300 |
| DVKSVYKYNN AEKSGFNKVL WVPRGKTIYV MSRKTDDPKG NDSGGADDIR FSSNVIKTFS | 360 |
| NRGYLDSVSL NGNTLSMRAW FWAGQSYKYQ FVFALDATTG KELSRKAANI ETRVDVKRAL | 420 |
| NNLTNSEKSG VKVQLAVPID KNIKIMIRRT NDPKGNERGG KSDYIFGGKT ISSYKSKFNQ | 480 |
| NSLSVAGTTL STRGWFWTQQ ATYKYQYIFV MDKNTNQELA RKLTPILNRS DVKNYLGNLK | 540 |
| GTNQTGFDTS IKLPGNKQAY IMVRRTNDPK GNGIGGYTEI SFPNKVVQTK ATLPVKPVKP | 600 |
| SQPAKPSQPV KPSQPAKPSQ PVKPSQPAKI DLTGTNDTQK AWFNALYASA QQLAKANDLF | 660 |
| PSVMMSQAIA ESAWGQSELA KTGNNLFGVK ADPSWTGAVV NRLTLENTTA ANQTVTGYRT | 720 |
| EAEGRAGKPA TTFVLANKGT PYYIYTGFRK YASQSESLRD YVTKIKTTVN GSTYRYQGAW | 780 |
| RSKAGSYQNA AHALKLGGYA TAPDYATNLT NRIEKYKLYV LD | 822 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 648 | 818 | 4.7e-28 | 0.9765625 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1705 | FlgJ | 2.90e-37 | 633 | 822 | 37 | 188 | Flagellum-specific peptidoglycan hydrolase FlgJ [Cell wall/membrane/envelope biogenesis, Cell motility]. |
| PRK08581 | PRK08581 | 5.42e-26 | 650 | 818 | 331 | 468 | amidase domain-containing protein. |
| PRK06347 | PRK06347 | 2.01e-24 | 587 | 822 | 90 | 303 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK05684 | flgJ | 9.34e-23 | 583 | 817 | 106 | 298 | flagellar assembly peptidoglycan hydrolase FlgJ. |
| smart00047 | LYZ2 | 4.91e-21 | 631 | 818 | 1 | 141 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDJ25195.1 | 0.0 | 25 | 822 | 1 | 798 |
| CEN29383.1 | 0.0 | 1 | 822 | 1 | 834 |
| QDJ28867.1 | 0.0 | 21 | 822 | 1 | 778 |
| QIW53050.1 | 4.72e-176 | 25 | 822 | 1 | 656 |
| QIW56712.1 | 5.29e-175 | 25 | 822 | 1 | 656 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4KNK_A | 4.01e-22 | 34 | 238 | 27 | 211 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNK_B Chain B, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_A Chain A, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_B Chain B, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_C Chain C, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_D Chain D, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 3LAT_A | 1.39e-19 | 75 | 238 | 52 | 202 | Crystalstructure of Staphylococcus peptidoglycan hydrolase AmiE [Staphylococcus epidermidis],3LAT_B Crystal structure of Staphylococcus peptidoglycan hydrolase AmiE [Staphylococcus epidermidis] |
| 3FI7_A | 2.33e-19 | 632 | 818 | 23 | 179 | CrystalStructure of the autolysin Auto (Lmo1076) from Listeria monocytogenes, catalytic domain [Listeria monocytogenes EGD-e] |
| 5T1Q_A | 5.78e-17 | 629 | 822 | 51 | 212 | ChainA, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_B Chain B, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_C Chain C, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_D Chain D, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 5DN5_A | 3.55e-09 | 663 | 817 | 27 | 149 | Structureof a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],5DN5_B Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],5DN5_C Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q2FZK7 | 3.31e-19 | 34 | 238 | 223 | 407 | Bifunctional autolysin OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=atl PE=1 SV=1 |
| Q99V41 | 5.70e-19 | 34 | 238 | 215 | 399 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
| Q931U5 | 5.70e-19 | 34 | 238 | 215 | 399 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=atl PE=1 SV=2 |
| Q6GAG0 | 5.71e-19 | 34 | 238 | 223 | 407 | Bifunctional autolysin OS=Staphylococcus aureus (strain MSSA476) OX=282459 GN=atl PE=3 SV=1 |
| Q5HH31 | 5.72e-19 | 34 | 238 | 223 | 407 | Bifunctional autolysin OS=Staphylococcus aureus (strain COL) OX=93062 GN=atl PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
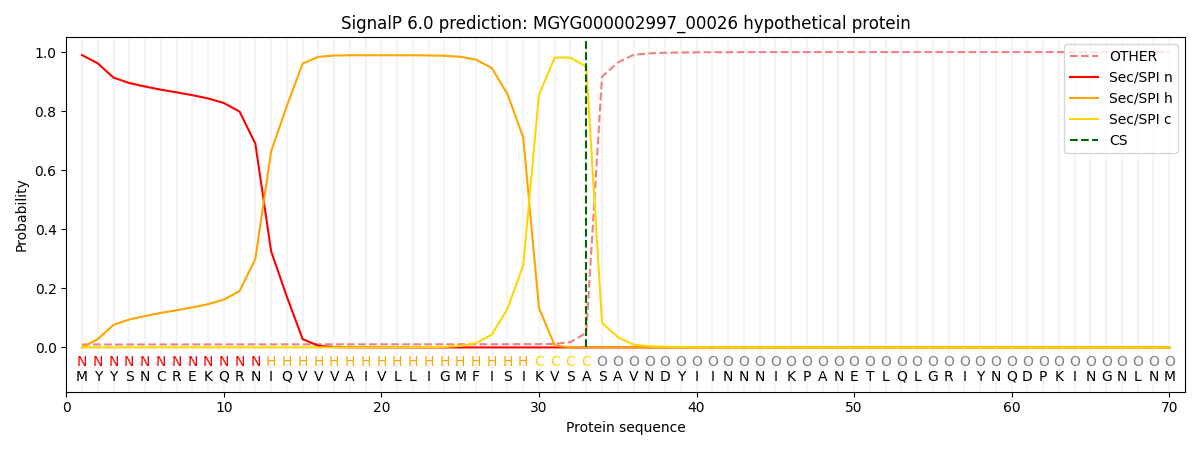
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.011701 | 0.987183 | 0.000436 | 0.000222 | 0.000209 | 0.000219 |