You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003832_00098
You are here: Home > Sequence: MGYG000003832_00098
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-1193 sp000436255 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RF39; UBA660; CAG-1193; CAG-1193 sp000436255 | |||||||||||
| CAZyme ID | MGYG000003832_00098 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 95131; End: 97767 Strand: - | |||||||||||
Full Sequence Download help
| MKKITFVLLF CFLFDFIMPF IDVFADVNYT FSSGSFHVAY ILDKNNTTSC NNIDTSTYDN | 60 |
| SNIEYVKSFN TYGEAFKYLN ELSDVDSKTL VIIGERKDKR GKYVNQILYS KYGLVDLNTT | 120 |
| GTTSTTTYVY TTATNNSVYT YINGHGYFGG VDAAFIDYNN GTSRAKIKMS GAIGWINSLL | 180 |
| SLNGSTYTGY DVVPLVSVKS PSYYYVTDDG ILVHRLSKKI TANNCYSTSM ALGKAPSYLE | 240 |
| SKDKNGNTIK YYSYDGNYFY TSIEDMTNDY REESVKRASN KVPYYNYYMY LPVRSKTNIS | 300 |
| SDTFDKYLES RNYTSKDKSA LYGEGLTFID AQNKYGVNAV ISFTTAIGES GWGTSDLAKT | 360 |
| KNNLFGHNAY DSSVMDSASG YKTVADGIYR HAYYYINAGF VETKDGIDRY HGAHLGNKGS | 420 |
| GINVKYASDP YWGEKIASQY YFLDNFADNV DYNKYTIGIK TSDKVVDVKS EPNTSSKTIY | 480 |
| KLKSASFNVS NIPVIVLGRV SGENINGSDI WYKIQSDALI DKSEERGDTV IRDATIDNIY | 540 |
| DWENDYVYVH SSYITLMDDS VNDIYTRKDG IFGLDNLTLD EKTNKVNIKG YLAIEGIDNS | 600 |
| LKNTFTYDLV LENENTSDVY ELPLNRITDS KNIPFTVSNN DKHDYTYSWF DGLLDFSEIK | 660 |
| EGNYTLYIRA RGKGYESKEL LSNVLALNIV NKYKDESGRG YEFKTNYYLK TIPIELFIRD | 720 |
| NGLISYESGP TSDNMFNQYQ NIELKDGNLK ISGTSFNVGG DYSTKKDVKR KIIFENINTF | 780 |
| KRYEFDLDYV DNGDYKITLV VPDNLDKTRA WFNKSIDLTS LEKGTYAIYI KTTSNVEDYG | 840 |
| ELIDIFAREL PKSNSYKDRN YSLKLNENAR FRIELIIK | 878 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.32e-17 | 285 | 456 | 67 | 242 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam01832 | Glucosaminidase | 1.29e-11 | 327 | 394 | 3 | 78 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| smart00047 | LYZ2 | 2.81e-07 | 327 | 441 | 15 | 140 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| COG1705 | FlgJ | 9.78e-05 | 314 | 366 | 32 | 92 | Flagellum-specific peptidoglycan hydrolase FlgJ [Cell wall/membrane/envelope biogenesis, Cell motility]. |
| NF038016 | sporang_Gsm | 1.75e-04 | 328 | 366 | 163 | 209 | sporangiospore maturation cell wall hydrolase GsmA. The peptidoglycan-hydrolyzing enzyme GsmA occurs in some sporangia-forming members of the Actinobacteria, such as Actinoplanes missouriensis, and is required for proper separation of spores. GsmA proteins have one or two SH3 domains N-terminal to the hydrolase domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIK69980.1 | 2.25e-78 | 128 | 557 | 125 | 537 |
| QNM11023.1 | 2.29e-75 | 148 | 562 | 163 | 553 |
| QSI26463.1 | 8.09e-73 | 111 | 555 | 91 | 505 |
| AYV34816.1 | 8.63e-70 | 113 | 602 | 100 | 568 |
| VEH84213.1 | 1.14e-68 | 82 | 552 | 72 | 519 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 3.08e-21 | 239 | 438 | 76 | 277 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 2.12e-19 | 239 | 438 | 444 | 645 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 2.34e-19 | 239 | 438 | 488 | 689 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
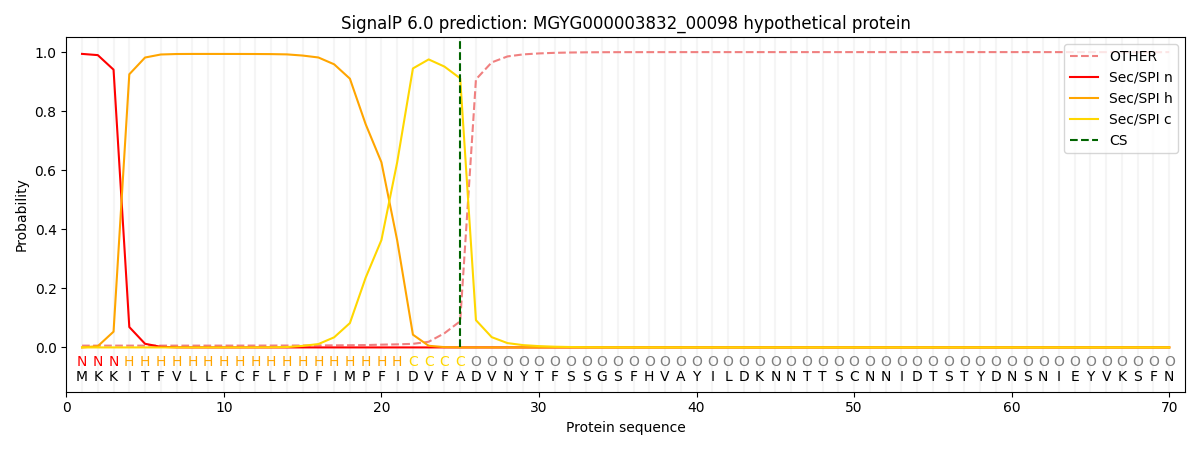
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.009532 | 0.989430 | 0.000286 | 0.000260 | 0.000210 | 0.000235 |

