You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001560_00072
You are here: Home > Sequence: MGYG000001560_00072
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
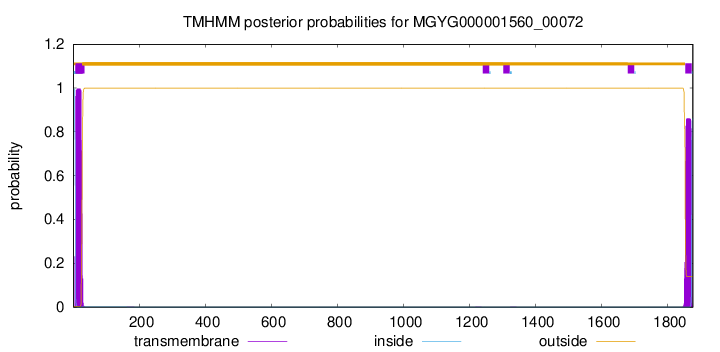
TMHMM annotations
Basic Information help
| Species | Massilioclostridium coli | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Massilioclostridium; Massilioclostridium coli | |||||||||||
| CAZyme ID | MGYG000001560_00072 | |||||||||||
| CAZy Family | CBM67 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 96897; End: 102527 Strand: + | |||||||||||
Full Sequence Download help
| MKKFYRSIAA ILAVCMLFTM AFTVSATDQT LYTMEILDLT VNNRTNPQGL DDDTPSFGWR | 60 |
| MDSNLVGQKQ TAYHVIVTEK FSGETMWDSG KVEGNVSQYI EYAGKELSPE SDYNWNVTVY | 120 |
| DSNGKAYLSD TATFTTGLMA DRLIPEEQPS IQDLTSYTID TDFVIHSGCM GLLFGVQDSN | 180 |
| NFYMWQINAG DLSINPHQWS NGGINTNFPH PSIKDALGVD SILETPLHLT MKMEEGTVTS | 240 |
| WINDVQIGEP LSIGQVKLGR IGFRQAVNES VDIDQITVKD QDGQVVFEED FSDQTSNPFD | 300 |
| GGTVENGQLK MNSTNLVFQK DTELENTTFT IDFDGQVVNT ALGPLIGGQD NSNFYMWQIN | 360 |
| AAERVFKPHQ WSGGNITTLK QVSLDPVFGE GQPIEGVPFH MTIQANNGEV KTYLNKIGEQ | 420 |
| PQLIDTIQLS PFSVGKFGFR QMKDHVSNEV GLIDNLKIVG ENGSILLEDD FSSPYNTTFP | 480 |
| SGSITEDGWF CAENGLFFQN ETEEVLGWSG AQWIGSPEIS VDSGKIPVYN LYYDLQLVEG | 540 |
| STKAAALVCG NDYRLMDKTK NNYRVEGESY LSFELDASNV SAGGSPVLNL YRKGFCSDDY | 600 |
| KNPDATTLMR SFSLDNIGVT AENLYTTPIR FSLTCYGGGF RIYVNDQNYG EVVINPTGGT | 660 |
| QDQPCYTYLN SIGFLTEPGQ QAEFQNYEIR TWDTAHAVMF DKTTGATYSI FEGQDGVSVQ | 720 |
| DHFIMVGDQA KQVKILAEPD YKSNQLLRKE FNADQPIQSA RLYITSRGIY EPYVNGERIG | 780 |
| EDWFNPGFTQ YNKTITYSTY DITDQLQQGT NAIGVRLASG WWHDEMTFDL SKYNYWGNKP | 840 |
| SLLAKVVVTY ADGTKDTIVT DESWSYNNTD SPIIYAGFFN GEVYDARKEA TIEGWNQPGY | 900 |
| TGENWYNASV IVPLPENANP HIIARVDEPV RHVETLSAKF DKEPDPNIYI YDMGTNMVGV | 960 |
| PEITFPEMPA GTEITIRYSE IYYPELDPSN PYYYGEMSGK LLTENLRAAL CTDTYIAKGT | 1020 |
| KGGETYRPSF TFHGYRYIEI SGLEEPLPEE NIKGIVLSSI EGTTSTYESS NELTNQLHKN | 1080 |
| IQRSLLGNHV SIPTDCPQRD ERMGWTGDAQ VFSRTATYYG DMNLLYEGWE GTIRDAQTAS | 1140 |
| GTIPQTAPNL GWGASVEVAW PAAIVIPTWE IYKQYGNTRV IESNLDAMKA FLDSIGNDKF | 1200 |
| APDSYLTKGT ALTEHLALVG TDSPLCSNVL YAYMLGLFSK MAAAIGEDEL AQHYGDLSVK | 1260 |
| VKEEWNQVFI NAATGQTQNS SGGIQDTQAS YALAIEYNVI SDENRDRAAE LLDQACERGY | 1320 |
| NNVPYTITTG FIGTAPLLPA LTDVGKVDTA YKMFEQTEFA SWLYPVTQGA TSIWERWNGY | 1380 |
| TIENGFSGLN YMNSFNHYAA GAVGAWMMND HVGITSDEEH PGFQQFILQP TLGGDFTYVN | 1440 |
| GGYDSIYGHI TSNWKVNQGK LTAYEAEVPA NTTATLYLPM DQNTVIGDLP EGVSFTGWDT | 1500 |
| RYGNSVAVFQ LVAGGYQFSV ENNGLTVALN DGYVTEGTPE PPVGDANKTI LDKVITEANR | 1560 |
| LKETEEYTNA IPAVKESFDK VLADAQAVYD NPSATQKEVD TAWMNLMEEI HKLGFQKGDK | 1620 |
| TALQELYYQV KDIDLSQYRD GAAKENFKTA LVNAETVLAD ENALQNEIDK AYNDLKSAYE | 1680 |
| ALEKLADKSQ LKALLDECAG YQKEEYTPVT WEVFEGIQKK AQEVYDNANA TQEEINAAVD | 1740 |
| ELLSGMLQLR FKADKSLLQN LIAELEEMDF SQYTEESVAA FQIVLAEAKT ILSDDNISIE | 1800 |
| DQQIVDQQVS NLKSAYKGLI GKQTDSTEFV DITPSNENSQ ETKPIEAANT GDVSPIAGLI | 1860 |
| LLGLAGITVA AFQKRK | 1876 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH78 | 944 | 1478 | 2.3e-142 | 0.9920634920634921 |
| CBM67 | 740 | 914 | 5.5e-34 | 0.9034090909090909 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17389 | Bac_rhamnosid6H | 8.44e-97 | 1064 | 1407 | 3 | 336 | Bacterial alpha-L-rhamnosidase 6 hairpin glycosidase domain. This family consists of bacterial rhamnosidase A and B enzymes. L-Rhamnose is abundant in biomass as a common constituent of glycolipids and glycosides, such as plant pigments, pectic polysaccharides, gums or biosurfactants. Some rhamnosides are important bioactive compounds. For example, terpenyl glycosides, the glycosidic precursor of aromatic terpenoids, act as important flavouring substances in grapes. Other rhamnosides act as cytotoxic rhamnosylated terpenoids, as signal substances in plants or play a role in the antigenicity of pathogenic bacteria. |
| pfam08531 | Bac_rhamnosid_N | 6.72e-60 | 756 | 934 | 2 | 172 | Alpha-L-rhamnosidase N-terminal domain. This family consists of bacterial rhamnosidase A and B enzymes. This domain is probably involved in substrate recognition. |
| pfam05592 | Bac_rhamnosid | 5.22e-28 | 942 | 1058 | 1 | 102 | Bacterial alpha-L-rhamnosidase concanavalin-like domain. This family consists of bacterial rhamnosidase A and B enzymes. L-Rhamnose is abundant in biomass as a common constituent of glycolipids and glycosides, such as plant pigments, pectic polysaccharides, gums or biosurfactants. Some rhamnosides are important bioactive compounds. For example, terpenyl glycosides, the glycosidic precursor of aromatic terpenoids, act as important flavouring substances in grapes. Other rhamnosides act as cytotoxic rhamnosylated terpenoids, as signal substances in plants or play a role in the antigenicity of pathogenic bacteria. |
| pfam17390 | Bac_rhamnosid_C | 6.80e-17 | 1421 | 1479 | 7 | 64 | Bacterial alpha-L-rhamnosidase C-terminal domain. This family consists of bacterial rhamnosidase A and B enzymes. L-Rhamnose is abundant in biomass as a common constituent of glycolipids and glycosides, such as plant pigments, pectic polysaccharides, gums or biosurfactants. Some rhamnosides are important bioactive compounds. For example, terpenyl glycosides, the glycosidic precursor of aromatic terpenoids, act as important flavouring substances in grapes. Other rhamnosides act as cytotoxic rhamnosylated terpenoids, as signal substances in plants or play a role in the antigenicity of pathogenic bacteria. |
| pfam06008 | Laminin_I | 6.58e-05 | 1580 | 1749 | 65 | 252 | Laminin Domain I. coiled-coil structure. It has been suggested that the domains I and II from laminin A, B1 and B2 may come together to form a triple helical coiled-coil structure. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGH33563.1 | 1.48e-306 | 486 | 1569 | 142 | 1230 |
| AIF65800.1 | 6.47e-233 | 485 | 1518 | 116 | 1159 |
| QGH37059.1 | 7.28e-230 | 485 | 1518 | 125 | 1180 |
| QOG04730.1 | 4.06e-229 | 499 | 1517 | 152 | 1192 |
| AOZ98905.1 | 2.67e-228 | 503 | 1523 | 140 | 1182 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3W5M_A | 6.34e-134 | 743 | 1521 | 302 | 1031 | CrystalStructure of Streptomyces avermitilis alpha-L-rhamnosidase [Streptomyces avermitilis MA-4680 = NBRC 14893],3W5N_A Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase complexed with L-rhamnose [Streptomyces avermitilis MA-4680 = NBRC 14893] |
| 6I60_A | 6.19e-109 | 746 | 1521 | 178 | 942 | Structureof alpha-L-rhamnosidase from Dictyoglumus thermophilum [Dictyoglomus thermophilum H-6-12],6I60_B Structure of alpha-L-rhamnosidase from Dictyoglumus thermophilum [Dictyoglomus thermophilum H-6-12] |
| 6GSZ_A | 1.53e-105 | 748 | 1479 | 135 | 847 | Crystalstructure of native alfa-L-rhamnosidase from Aspergillus terreus [Aspergillus terreus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q82PP4 | 3.51e-133 | 743 | 1520 | 302 | 1030 | Alpha-L-rhamnosidase OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=SAVERM_828 PE=1 SV=1 |
| T2KNB2 | 2.57e-119 | 727 | 1483 | 162 | 883 | Alpha-L-rhamnosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22090 PE=1 SV=2 |
| P9WF03 | 6.24e-104 | 736 | 1520 | 167 | 910 | Alpha-L-rhamnosidase OS=Alteromonas sp. (strain LOR) OX=1537994 GN=LOR_34 PE=1 SV=1 |
| T2KPL4 | 5.49e-59 | 748 | 1478 | 188 | 906 | Alpha-L-rhamnosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22170 PE=2 SV=1 |
| E8MGH9 | 4.77e-08 | 1687 | 1819 | 1667 | 1803 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
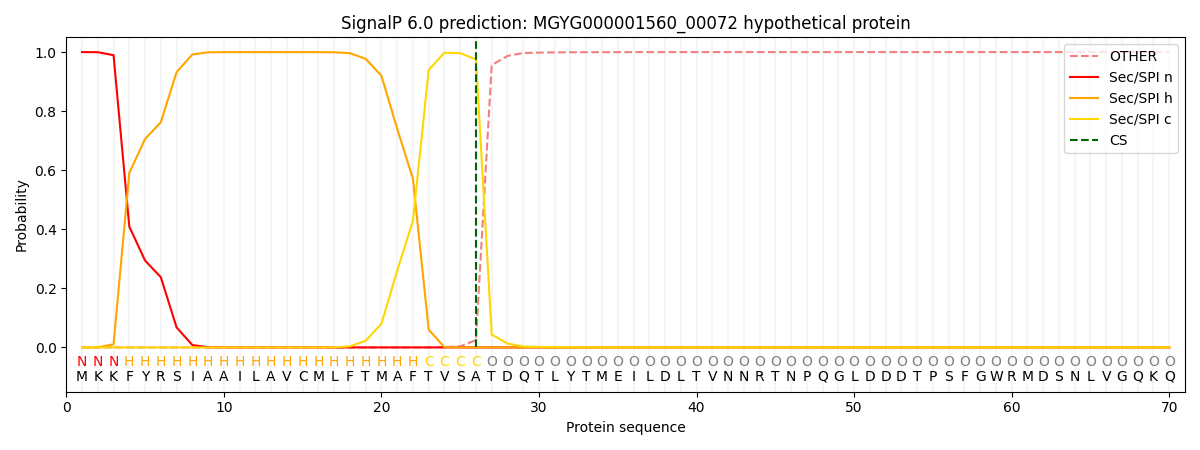
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000354 | 0.998771 | 0.000254 | 0.000207 | 0.000199 | 0.000182 |