You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001289_00038
You are here: Home > Sequence: MGYG000001289_00038
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
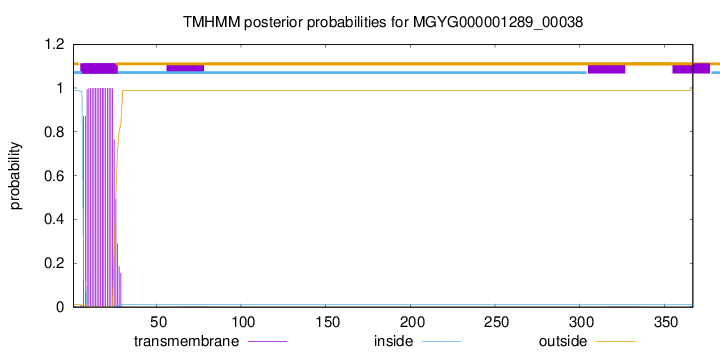
TMHMM annotations
Basic Information help
| Species | Streptococcus sp900543065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus; Streptococcus sp900543065 | |||||||||||
| CAZyme ID | MGYG000001289_00038 | |||||||||||
| CAZy Family | GH8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 38296; End: 39399 Strand: + | |||||||||||
Full Sequence Download help
| MKTKKMRYVW FLAILCIFCL TLFLARTRSK VDMRNRIYRQ WQDHFVVSKG KESYVQTNNS | 60 |
| KDQTVVLSEA QSYGMVITVA AAKKGQAQQE DFERLYQYYL NHRITDTQLM SWKQTIVDGK | 120 |
| ASAEDHNATD GDLYIAYSLL EAAQQWPKQA KKYQDQAKAI LEDVLKYNYN EETGVLTVGN | 180 |
| WANQDSNFYY LMRTSDTLPA QFQLFYKVTN DSKWLDIKDK MLHQLDVVSS KTSSGLLPDF | 240 |
| IWVDHDGASV ADPNTIESKY DGSYSYNACR LPYNLSQSKD KTSQKLLKKM LGFFKKQRNL | 300 |
| YAGYDLKGKA LNDHQAASFL APIVYASESG EGYLKLVQQN KYIFTQDLPL NNYYDATMTT | 360 |
| MIALELF | 367 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3405 | BcsZ | 6.39e-20 | 5 | 314 | 1 | 296 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
| pfam01270 | Glyco_hydro_8 | 5.31e-18 | 38 | 364 | 8 | 317 | Glycosyl hydrolases family 8. |
| PRK11097 | PRK11097 | 2.15e-09 | 51 | 270 | 32 | 248 | cellulase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUB39088.1 | 1.66e-269 | 1 | 367 | 1 | 367 |
| AEH56684.1 | 4.27e-224 | 1 | 367 | 1 | 367 |
| AFJ25304.1 | 1.43e-222 | 1 | 367 | 1 | 367 |
| AGY40742.1 | 2.42e-200 | 1 | 367 | 1 | 367 |
| AGY37425.1 | 2.42e-200 | 1 | 367 | 1 | 367 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5XD0_A | 2.21e-33 | 27 | 322 | 54 | 359 | ApoStructure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 [Paenibacillus sp. X4],5XD0_B Apo Structure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 [Paenibacillus sp. X4] |
| 1V5C_A | 6.59e-22 | 68 | 337 | 73 | 350 | Thecrystal structure of the inactive form chitosanase from Bacillus sp. K17 at pH3.7 [Bacillus sp. (in: Bacteria)],1V5D_A The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 [Bacillus sp. (in: Bacteria)],1V5D_B The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 [Bacillus sp. (in: Bacteria)] |
| 7CJU_A | 7.26e-22 | 68 | 337 | 79 | 356 | Crystalstructure of inactive form of chitosanase crystallized by ammonium sulfate [Bacillus sp. K17-2],7CJU_B Crystal structure of inactive form of chitosanase crystallized by ammonium sulfate [Bacillus sp. K17-2],7XGQ_A Chain A, chitosanase [Bacillus sp. K17-2],7XGQ_B Chain B, chitosanase [Bacillus sp. K17-2] |
| 6VC5_A | 1.34e-09 | 35 | 361 | 2 | 308 | 1.6Angstrom Resolution Crystal Structure of endoglucanase from Komagataeibacter sucrofermentans [Komagataeibacter sucrofermentans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P19254 | 1.21e-32 | 27 | 322 | 54 | 359 | Beta-glucanase OS=Niallia circulans OX=1397 GN=bgc PE=3 SV=1 |
| P29019 | 5.75e-22 | 68 | 326 | 129 | 395 | Endoglucanase OS=Bacillus sp. (strain KSM-330) OX=72575 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
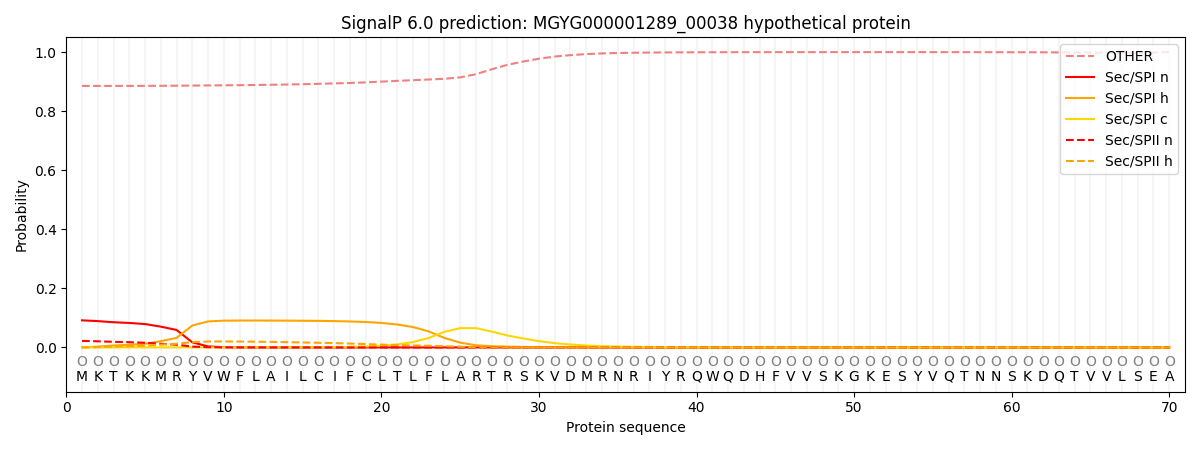
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.890290 | 0.085081 | 0.022760 | 0.000305 | 0.000210 | 0.001352 |