You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002579_00311
You are here: Home > Sequence: MGYG000002579_00311
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
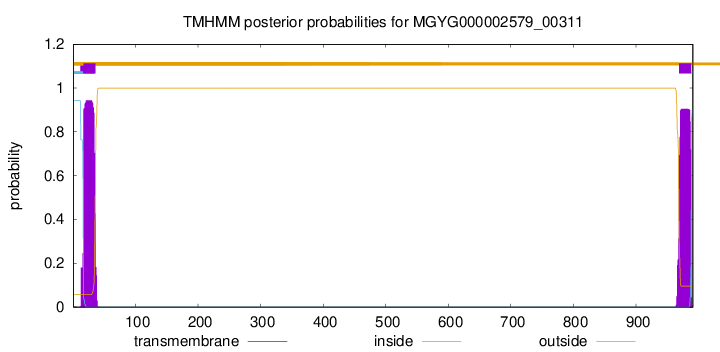
TMHMM annotations
Basic Information help
| Species | Ligilactobacillus sp900765635 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Ligilactobacillus; Ligilactobacillus sp900765635 | |||||||||||
| CAZyme ID | MGYG000002579_00311 | |||||||||||
| CAZy Family | GH84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 39296; End: 42271 Strand: - | |||||||||||
Full Sequence Download help
| MQNKKTHYKM YKSGKKWVFM GITTVSLVTV LGATTHPAKA DNVNHSTPSV LATKNTDQTV | 60 |
| SIQSSLDAVN KELANKIIVL LKENNISFNQ ISIGTADDQT TSDLVGQLNG NSVNNLKDNG | 120 |
| YLIKSGTINN QTALAIQGKD ATGLFYALNQ VITTINDKQS VTNLAVYESP QMSIRGVIEG | 180 |
| FYGQPWSNQA RKDMFKFMGQ HRMNTYIYSP KDDTYLRENW RKPYPQDKLN EIKELVDEAK | 240 |
| KNHVEFVYAL SPGNDITYSS TADYQATIAK FDQLRSIGVS QFYIALDDIN PVINDTDAAV | 300 |
| FPAHDTPNYP NNSWSPLADA QAYYLNKVQR EYVEKNNLPA LWLVPTNYSG SAQDPFKEAQ | 360 |
| GLALDKDIHI QWTGEGVFSD KITADSINKA KETYHTDKLF IWDNFPVNDS NQDRLYLNPV | 420 |
| VGRADDLYQT TEGFTSNPMI EPYASWIGIG SYADYMWNSS TYNPQTSLNN VLAEIAGNDS | 480 |
| TVLDSLKAFA DLNQYWDHGS ADEQTKAPIL SSLVTAYQKS SPGSDLHAQA LANLKAQLTK | 540 |
| IAQASVTLQH LAIPGFYNDA LPWINAASHW AKASLANIEI TEALQSANPV NLDTLGSNLV | 600 |
| TLKSEIAQAK VKAIVDGRTD SSEPAIVPSV GDGVFETLTN LNGDFNNLDS WFGFTPLKVT | 660 |
| NNKYTGTAST QIPVYQNNGV ANITDDSDTS LFWSSRNTAK GDTITLKLTK PQQIASIVLK | 720 |
| QGDTDTATSG DMFTDATVYI GNKEDGSDKQ AVGHIASSGL AQVDLSTPVT GQYVFIVANS | 780 |
| DSSSWLKIRD LSVYGTSNFK LTNSSSSDGK SIQTLFDGSL TTDFTAKIND TSKSATIEQV | 840 |
| SDTPVTNANS LVLVGKAKGT LYVRSNGTWQ KVGKTDGSKQ INEFGLTNSQ SKNKAATGFD | 900 |
| GIRLVLDPGS QNVVLSELNF STKTAPVYSE DTNKNPETPS DKPTNTNKNT NKNTNTKKDG | 960 |
| NLPQTGEQVS ILASLAGLFL AGLAVLGFKK H | 991 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 175 | 490 | 7e-99 | 0.9932203389830508 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 2.39e-140 | 175 | 489 | 1 | 293 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| TIGR03715 | KxYKxGKxW | 2.61e-09 | 5 | 27 | 1 | 23 | KxYKxGKxW signal peptide. This model describes a novel form of signal peptide that occurs as an N-terminal domain with a recognizable motif, reminiscent of the YSIRK and PEP-CTERM forms of signal peptide. This domain tends to occur on long, low-complexity (usually Serine-rich and heavily glycosylated) proteins of the Firmicutes, and (as with YSIRK) the majority of these proteins have the LPXTG cell wall-anchoring motif at the C-terminus. |
| pfam00754 | F5_F8_type_C | 2.50e-05 | 667 | 785 | 3 | 119 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| TIGR01167 | LPXTG_anchor | 3.43e-04 | 962 | 990 | 2 | 30 | LPXTG-motif cell wall anchor domain. This model describes the LPXTG motif-containing region found at the C-terminus of many surface proteins of Streptococcus and Streptomyces species. Cleavage between the Thr and Gly by sortase or a related enzyme leads to covalent anchoring at the new C-terminal Thr to the cell wall. Hits that do not lie at the C-terminus or are not found in Gram-positive bacteria are probably false-positive. A common feature of this proteins containing this domain appears to be a high proportion of charged and zwitterionic residues immediatedly upstream of the LPXTG motif. This model differs from other descriptions of the LPXTG region by including a portion of that upstream charged region. [Cell envelope, Other] |
| pfam00746 | Gram_pos_anchor | 0.003 | 962 | 991 | 9 | 39 | LPXTG cell wall anchor motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| APU72174.1 | 1.66e-249 | 81 | 733 | 143 | 792 |
| QGH68398.1 | 7.31e-184 | 91 | 918 | 117 | 912 |
| AER06669.1 | 8.07e-181 | 68 | 918 | 63 | 886 |
| ALD69353.1 | 1.84e-180 | 68 | 918 | 92 | 915 |
| QJW35068.1 | 3.68e-175 | 93 | 918 | 111 | 910 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PV5_A | 1.72e-65 | 91 | 582 | 100 | 576 | Structureof CpGH84B [Clostridium perfringens ATCC 13124] |
| 2CBI_A | 2.02e-63 | 63 | 612 | 46 | 584 | Structureof the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase [Clostridium perfringens],2CBI_B Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase [Clostridium perfringens],2CBJ_A Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase in complex with PUGNAc [Clostridium perfringens],2CBJ_B Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase in complex with PUGNAc [Clostridium perfringens],2V5C_A Family 84 glycoside hydrolase from Clostridium perfringens, 2.1 Angstrom structure [Clostridium perfringens],2V5C_B Family 84 glycoside hydrolase from Clostridium perfringens, 2.1 Angstrom structure [Clostridium perfringens],2VUR_A Chemical dissection of the link between Streptozotocin, O-GlcNAc and pancreatic cell death [Clostridium perfringens],2VUR_B Chemical dissection of the link between Streptozotocin, O-GlcNAc and pancreatic cell death [Clostridium perfringens],2X0Y_A Screening-based discovery of drug-like O-GlcNAcase inhibitor scaffolds [Clostridium perfringens],2X0Y_B Screening-based discovery of drug-like O-GlcNAcase inhibitor scaffolds [Clostridium perfringens] |
| 5OXD_A | 3.47e-63 | 63 | 612 | 48 | 586 | Complexof a C. perfringens O-GlcNAcase with a fragment hit [Clostridium perfringens] |
| 2J62_A | 3.77e-63 | 63 | 612 | 46 | 584 | Structureof a bacterial O-glcnacase in complex with glcnacstatin [Clostridium perfringens],2J62_B Structure of a bacterial O-glcnacase in complex with glcnacstatin [Clostridium perfringens],2WB5_A GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation [Clostridium perfringens],2WB5_B GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation [Clostridium perfringens] |
| 4ZXL_A | 1.32e-62 | 63 | 612 | 38 | 576 | CpOGAD298N in complex with Drosophila HCF -derived Thr-O-GlcNAc peptide [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8XL08 | 4.48e-61 | 63 | 612 | 76 | 614 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| Q0TR53 | 2.64e-60 | 63 | 612 | 76 | 614 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q89ZI2 | 1.77e-51 | 130 | 463 | 107 | 419 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
| O60502 | 1.82e-38 | 176 | 512 | 63 | 375 | Protein O-GlcNAcase OS=Homo sapiens OX=9606 GN=OGA PE=1 SV=2 |
| Q8VIJ5 | 1.82e-38 | 176 | 512 | 63 | 375 | Protein O-GlcNAcase OS=Rattus norvegicus OX=10116 GN=Oga PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
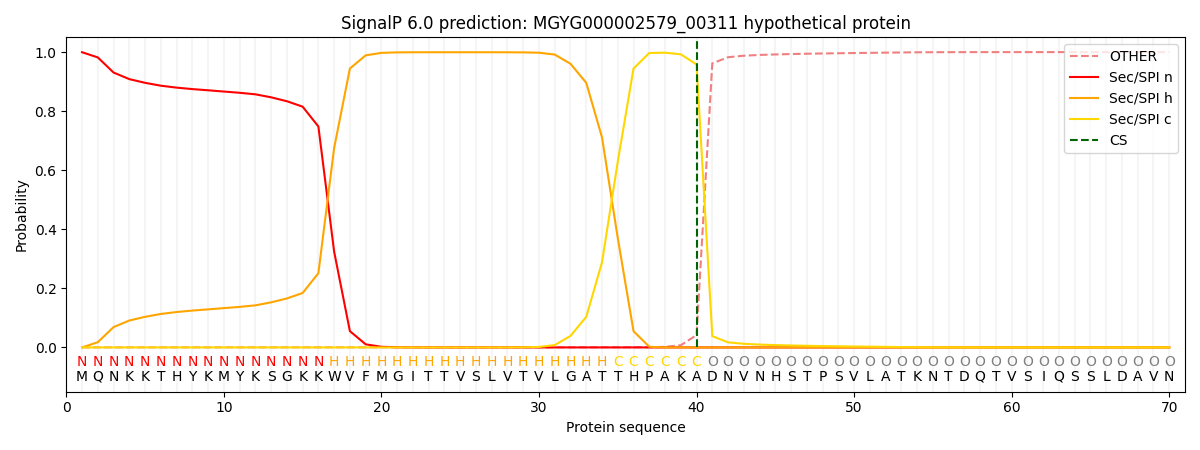
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000868 | 0.998170 | 0.000374 | 0.000226 | 0.000173 | 0.000155 |