You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003768_00203
You are here: Home > Sequence: MGYG000003768_00203
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
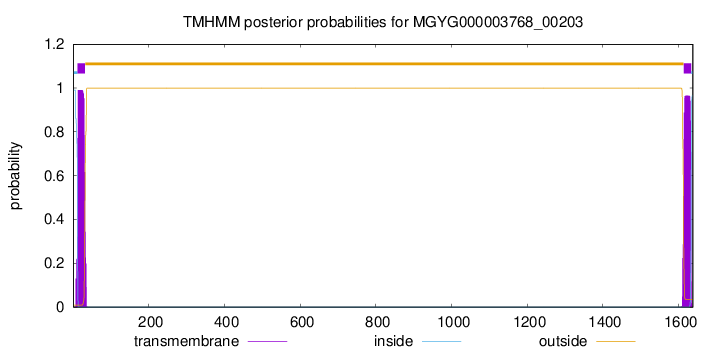
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; Clostridium_A; | |||||||||||
| CAZyme ID | MGYG000003768_00203 | |||||||||||
| CAZy Family | GH85 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 39915; End: 44831 Strand: - | |||||||||||
Full Sequence Download help
| MKKRRKWRRS VISFMLASAM VLPGVGTAFA GGDTLPPREG EQGYKGEHQP STHGYNRANI | 60 |
| LNWSPETDGY AEFMRSKVPL QERNEAFTAT QANPLLDQEV ESLSLMGDYG IGFFDSFQYN | 120 |
| DQFSQYLFNF WQYLDYHASW HGMATDPQPH DLFTPGNPEG QGKFEFGVVN LPNPAYTNAA | 180 |
| HKNGVKSLGC IFFPRTEHTE DFVYQDENGR FPIADKLVEI AKYYGFDGYF INAEEDLKPE | 240 |
| QMPIYEEFCR AMTSQGIYIQ AYASCLYGPD NEDKWGTMDY GEKNAGKFSN WLKGADDETI | 300 |
| AANSLYMNPD PTKQQIDDSV TSMEALGLDP KKVVFQTLEA GQTGFSGKRG TLFNTLDENL | 360 |
| VPRTGLAFLG ADTSKNHLDE QVFGHVGKNS YKYNKRDNPE YQKYVFARER TWWSGALDQP | 420 |
| YYEPFKTQQP DDYFPEELCQ EILDATPDPY QTANNPKRGQ ANEGEDYQSW PGLAAFISER | 480 |
| SVIDGSNFYT NFNTGHGMQY FVDGQVSNDN EWSNINIQDI LPTWQWWIET EGTRLDVDFD | 540 |
| YGEKYSPAFE LNQVGGYNGG SSLVVKGKLD AENTLRLYKT NLEVKENTKI SVTYNKTSAT | 600 |
| DNSKMELAVI FKNDPNTVVT FNVPQANQET DGWVTKDISL GDYAGEEIVS LGFNFDPGKE | 660 |
| TMEDYQMNIG ELKVSDGSVA APSVPTGLSI DESFETSETY ISWDLDDYDA VQRYNVYMKD | 720 |
| GDGNRTYLGG TYDDIYYIKD LYDVQGDITI QVTAVGKDGT ESAPAEVVLQ QDKAVRDIQV | 780 |
| TQEAGKFDIS WTNPETDYDH AEVTVTFESH PDDVYTAEAA KGETGINLEI PIMDGTNYTV | 840 |
| RVSLMDAEGQ VIGFMDQTGQ LEDYYCHPYD QDVRFANNLL SFAPPSVNDW STMYAFYKGE | 900 |
| PIKFRSGNIG AIRGYDDMRG ILINDDCGVV EVELEDYAGN RSEQIRVPFH KDGMTCAAID | 960 |
| ETAFPDPALL EAVKKVAETP ADLLNVTTLD LSNSEVKDLT GLRYLRNLES LNLSNCTSLE | 1020 |
| LIQEQDLSLN VHLKEINLTG CTQLKAASLA DTTLETIVCE DVSALSQLYF LDVSGSRFDL | 1080 |
| SEGTPEREFV DAVTAQAEGK GDFTVNLPNP VNLAYNMKAT DTNLIMASYL FDGDTSNPMR | 1140 |
| CVWIPNIPSH VTLQFDSPKE VLSYHMDVPN DDPSVGLKDF RLQGSMDGQQ WTDLSVVTGH | 1200 |
| TGESCTITVE NPQAYTYYRF QIDGKVPASG PAKLTELEMT GYGDVTFTAG VQYDNQRPVA | 1260 |
| YEKEMPETVR LEKQLGGTYD LTANLENNVT IHGNDLASLK GADFIDQDYD LDKVAQNKVQ | 1320 |
| YIRVTGEDGS QLTNEISLDQ DGAYTVEYLT YKDAGYQART VATQTVYVRG VTTVLEKIIA | 1380 |
| EAEELLNNGA LDNTMEVVVE EFKAALAHAK EILAKDDATQ QEINDATKRL LDVMAKVDWK | 1440 |
| QGDKTALQVA VDIANTIKPD LDLYVEEGKQ EFLDALAKGE ELLASGNAWA DEIQTATEEL | 1500 |
| IKAMSNLRMA PNKDILNDMI DKASGLDLSD YTQDSAAVLT SALAEAQEVA ADENATQEEV | 1560 |
| DEAADTLEAA MNGLVFVNSD ENNNTQDDTI SNPDDTENAP VGDGTAPIKT GDAGAAGLAG | 1620 |
| LAVMSALGIV WMLRKREK | 1638 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH85 | 106 | 501 | 7.2e-61 | 0.9936507936507937 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4724 | COG4724 | 1.13e-78 | 44 | 676 | 23 | 551 | Endo-beta-N-acetylglucosaminidase D [Carbohydrate transport and metabolism]. |
| cd06547 | GH85_ENGase | 5.29e-55 | 102 | 532 | 3 | 339 | Endo-beta-N-acetylglucosaminidase (ENGase) hydrolyzes the N-N'-diacetylchitobiosyl core of N-glycosylproteins. The beta-1,4-glycosyl bond located between two N-acetylglucosamine residues is hydrolyzed such that N-acetylglucosamine 1 remains with the protein and N-acetylglucosamine 2 forms the reducing end of the released glycan. ENGase is a key enzyme in the processing of free oligosaccharides in the cytosol of eukaryotes. Oligosaccharides formed in the lumen of the endoplasmic reticulum are transported into the cytosol where they are catabolized by cytosolic ENGases and other enzymes, possibly to maximize the reutilization of the component sugars. ENGases have an eight-stranded alpha/beta barrel topology and are classified as a family 85 glycosyl hydrolase (GH85) domain. The GH85 ENGases are sequence-similar to the family 18 glycosyl hydrolases, also known as GH18 chitinases. An ENGase-like protein is also found in bacteria and is included in this alignment model. |
| pfam03644 | Glyco_hydro_85 | 6.97e-52 | 121 | 497 | 3 | 291 | Glycosyl hydrolase family 85. Family of endo-beta-N-acetylglucosaminidases. These enzymes work on a broad spectrum of substrates. |
| cd21340 | PPP1R42 | 4.44e-06 | 960 | 1045 | 105 | 185 | protein phosphatase 1 regulatory subunit 42. Protein phosphatase 1 regulatory subunit 42 (PPP1R42), also known as leucine-rich repeat-containing protein 67 (lrrc67) or testis leucine-rich repeat (TLRR) protein, plays a role in centrosome separation. PPP1R42 has been shown to interact with the well-conserved signaling protein phosphatase-1 (PP1) and thereby increasing PP1's activity, which counters centrosome separation. Inhibition of PPP1R42 expression increases the number of centrosomes per cell while its depletion reduces the activity of PP1 leading to activation of NEK2, the kinase responsible for phosphorylation of centrosomal linker proteins promoting centrosome separation. |
| pfam07554 | FIVAR | 8.77e-06 | 1516 | 1575 | 3 | 69 | FIVAR domain. This domain is found in a wide variety of contexts, but mostly occurring in cell wall associated proteins. A lack of conserved catalytic residues suggests that it is a binding domain. From context, possible substrates are hyaluronate or fibronectin (personal obs: C Yeats). This is further evidenced by. Possibly the exact substrate is N-acetyl glucosamine. Finding it in the same protein as pfam05089 further supports this proposal. It is found in the C-terminal part of Bacillus sp. Gellan lyase, which is removed during maturation. Some of the proteins it is found in are involved in methicillin resistance. The name FIVAR derives from Found In Various Architectures. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUH30609.1 | 0.0 | 8 | 949 | 3 | 958 |
| SMF77083.1 | 9.03e-224 | 1 | 943 | 1 | 916 |
| AWB44945.1 | 5.33e-214 | 12 | 1210 | 21 | 1198 |
| QNO18499.1 | 5.01e-210 | 10 | 948 | 20 | 919 |
| ALA43266.1 | 5.20e-208 | 6 | 942 | 12 | 910 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2W91_A | 5.58e-57 | 121 | 777 | 78 | 640 | Structureof a Streptococcus pneumoniae family 85 glycoside hydrolase, Endo-D. [Streptococcus pneumoniae TIGR4],2W92_A Structure of a Streptococcus pneumoniae family 85 glycoside hydrolase, Endo-D, in complex with NAG-thiazoline. [Streptococcus pneumoniae TIGR4] |
| 3GDB_A | 2.02e-55 | 121 | 777 | 229 | 791 | Crystalstructure of Spr0440 glycoside hydrolase domain, Endo-D from Streptococcus pneumoniae R6 [Streptococcus pneumoniae R6] |
| 3FHA_A | 1.33e-43 | 60 | 757 | 16 | 604 | ChainA, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae],3FHA_B Chain B, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae],3FHA_C Chain C, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae],3FHA_D Chain D, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae] |
| 3FHQ_A | 1.79e-43 | 60 | 757 | 16 | 604 | ChainA, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae],3FHQ_B Chain B, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae],3FHQ_D Chain D, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae],3FHQ_F Chain F, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae] |
| 2VTF_A | 1.91e-43 | 60 | 757 | 21 | 609 | X-raycrystal structure of the Endo-beta-N-acetylglucosaminidase from Arthrobacter protophormiae E173Q mutant reveals a TIM barrel catalytic domain and two ancillary domains [Glutamicibacter protophormiae],2VTF_B X-ray crystal structure of the Endo-beta-N-acetylglucosaminidase from Arthrobacter protophormiae E173Q mutant reveals a TIM barrel catalytic domain and two ancillary domains [Glutamicibacter protophormiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9SRL4 | 3.26e-11 | 164 | 760 | 109 | 679 | Cytosolic endo-beta-N-acetylglucosaminidase 2 OS=Arabidopsis thaliana OX=3702 GN=ENGASE2 PE=1 SV=1 |
| A1L251 | 2.66e-07 | 164 | 255 | 142 | 235 | Cytosolic endo-beta-N-acetylglucosaminidase OS=Danio rerio OX=7955 GN=engase PE=2 SV=1 |
| Q8BX80 | 2.70e-07 | 164 | 255 | 156 | 250 | Cytosolic endo-beta-N-acetylglucosaminidase OS=Mus musculus OX=10090 GN=Engase PE=1 SV=1 |
| Q8NFI3 | 1.39e-06 | 164 | 255 | 164 | 258 | Cytosolic endo-beta-N-acetylglucosaminidase OS=Homo sapiens OX=9606 GN=ENGASE PE=1 SV=1 |
| P0C7A1 | 2.37e-06 | 60 | 255 | 91 | 256 | Cytosolic endo-beta-N-acetylglucosaminidase OS=Gallus gallus OX=9031 GN=ENGASE PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
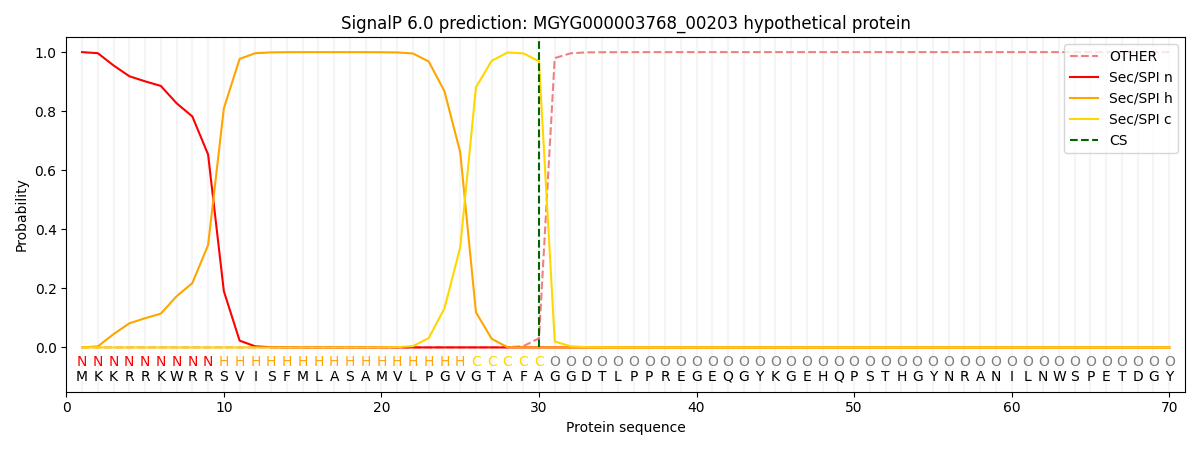
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000496 | 0.998488 | 0.000302 | 0.000319 | 0.000196 | 0.000165 |