You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000018_00340
You are here: Home > Sequence: MGYG000000018_00340
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
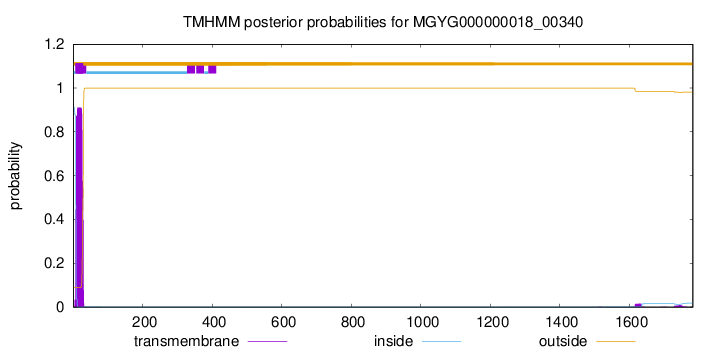
TMHMM annotations
Basic Information help
| Species | Coprococcus eutactus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Coprococcus; Coprococcus eutactus | |||||||||||
| CAZyme ID | MGYG000000018_00340 | |||||||||||
| CAZy Family | GH9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 373001; End: 378352 Strand: - | |||||||||||
Full Sequence Download help
| MRKGTGSRIL AYVLTLCMII GSITWPEITA KAETITKDLK PDTGWKTVTA ATDEWSDYGK | 60 |
| AEIRFSPSSD LASMKAIADA GYKTLKITYA VDTFTAASGQ NAGVMPFASY GSSWSNNDKW | 120 |
| IDLSKTGQFE TVLDLASIST TSTEKVAFGI QVANLQENST IKFRIVSAVL SGTKSTSGGS | 180 |
| SGESGGSGDS GSGSADLDSI GNTSSSVTAS LADGDGTAKG DGYYETEITI NNKSNSYIAD | 240 |
| WIAVADVSGS VTAVNDYSSW SDLKGVFSDG KLYIYPNTSK KSGAVNAGSS VRYSKLGYTG | 300 |
| TANGVSITGV KVYYSSQSGA FDSFIGSLSS SSGGAGDNTG EINTDVEYNY AKLLQESLYL | 360 |
| YDANMCGSDV SAKSEFSWRS NCHTEDAKTT YNGKTVDVSG GYHDAGDHAK FGLPQAYSAT | 420 |
| VLGLAHMEFA EAFADTATEA HYKRIMDRFV DYFKRCTVLG SDGSVQAFCY QVGDGNVDHG | 480 |
| YWGAPEKQSS RSGQATFTSD SDTCTDIVSE TAAALAAYYI NYKDKKALSY AEKLFTYADT | 540 |
| KAKKNSSGPA SGFYNSDSWE DDYALAAALL YKATGKSAYA TKYNNVYGGR TNPNWALCWN | 600 |
| NVAQAALLYS PNSSKKSVFV ENQSGLIASK TQSGDNNFCL IDSWGSARYN TAHQMTGLLY | 660 |
| DTIYGKNDYS SWANGQMKYI LGNNAGSKCF VVGYNKYSSK YPHHRASSGY QGSVTGNAYT | 720 |
| KQAHVLVGAL VGGPASSSTS YVDSSEDYNQ NEVALDYNAS LVGAAAGLYL YVKNSGTDEE | 780 |
| KAAQKVVPKS EVSSELRTIS GESGGNVTTE TTTKTTTTEK ATKNPSSGSS AGSTGSTTGS | 840 |
| STEKTTESST ETQVIPVKGI TFDQTALTMK VGESGQIKAT VTPADATDSS LVWTSSDRTK | 900 |
| VSVQNGKITA LAAGTATITA KAKDGSGVKA ECKVKVLEPG KLSCETSGKA WNALVYGYGD | 960 |
| VSSERIGLSN CGETELSDVK ASLKTGSNFQ ITVYPAGQIA AGQETSVEVR PVTGLAAGSY | 1020 |
| SDTLVITTAN GTANILLKAK VAKSENTANV SLAKKAVSSS SVTVESYVSG TNTGVEYAIS | 1080 |
| TVPVEDADSL TWQDGAYFAG CKAFTKYYVY GRMKATSNLN AGAMSKALEI VTLVSDPYTI | 1140 |
| DVGRLGDSEY VGALVDGNGN PTVRVSESGG NILVSFTQSG DYTVTGDGAD VAVDTGKAGS | 1200 |
| ITIDCAVVKK LTVDPGNSGT FVISVLGNNI VSDGIKCMED SAGSGVVKIN GNGDSSVIVS | 1260 |
| STPDAPAVKA DGDVEISGIK IKSEGKGVES AGTVKISGGS NKIEAVSEAV AAKDVEMTGG | 1320 |
| LLDATSTALG DDESVISADN SIKLVGGKIT ADASGSSTGG SFGVRSDDGK IIVDGDAVIG | 1380 |
| GAPTYSKDPV NSTGESIVMV KVTFVDENDG QICVSSFNKG SILDISKLDI TTKDGVAYSA | 1440 |
| SKPGYSLAWT DQTGKTYAAD ALYGAVDGDI TLKAVWTWIT VDISKSAKVI YKTTQKASYK | 1500 |
| ATYTGAKITP AVVVSVGNTT LVSGTDYTVT YSSNINAGTA KVVVKGKEKY KGTVTLTFAI | 1560 |
| AKRDIKKAKV AVSAKVLYTG KAVKPNTKVV YGKTKLTLNK NYKITCYSNK NFGKAKVIIT | 1620 |
| GIGNYGGSVV RYFNIVTKAG KVYTYGNYRY KITNASTSGR GTVTLVSAVK KTTSVTVPDA | 1680 |
| IKLGGKTFKV TAIGGGAFKG NTKLAKVVIS KNVKTIGSKA FYGCKNLKAL VVRNTAMTTK | 1740 |
| TLGAAAFTGT YAKMTVKVPA SKLAYYKKLL VARGVSKKAV IKK | 1783 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 349 | 766 | 1.2e-101 | 0.9976076555023924 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00759 | Glyco_hydro_9 | 3.75e-89 | 352 | 765 | 1 | 374 | Glycosyl hydrolase family 9. |
| PLN02345 | PLN02345 | 1.98e-35 | 372 | 769 | 18 | 459 | endoglucanase |
| PLN02420 | PLN02420 | 4.89e-35 | 348 | 780 | 40 | 517 | endoglucanase |
| PLN02340 | PLN02340 | 1.25e-34 | 348 | 796 | 29 | 522 | endoglucanase |
| PLN02613 | PLN02613 | 1.16e-32 | 347 | 768 | 24 | 478 | endoglucanase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK83841.1 | 0.0 | 1 | 1783 | 1 | 1782 |
| AFK82697.1 | 0.0 | 1 | 810 | 1 | 810 |
| QWT52133.1 | 7.88e-121 | 345 | 769 | 33 | 455 |
| ADD61854.1 | 5.31e-120 | 335 | 936 | 179 | 730 |
| QNL98526.1 | 3.35e-116 | 335 | 936 | 198 | 749 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2YIK_A | 3.45e-100 | 338 | 806 | 28 | 553 | ChainA, Endoglucanase [Acetivibrio thermocellus] |
| 1IA6_A | 4.32e-73 | 348 | 767 | 4 | 425 | CrystalStructure Of The Cellulase Cel9m Of C. Cellulolyticum [Ruminiclostridium cellulolyticum],1IA7_A Crystal Structure Of The Cellulase Cel9m Of C. Cellulolyticium In Complex With Cellobiose [Ruminiclostridium cellulolyticum] |
| 2XFG_A | 8.32e-55 | 348 | 769 | 24 | 460 | ChainA, ENDOGLUCANASE 1 [Acetivibrio thermocellus] |
| 4DOD_A | 3.88e-53 | 348 | 769 | 26 | 460 | Thestructure of Cbescii CelA GH9 module [Caldicellulosiruptor bescii],4DOE_A The liganded structure of Cbescii CelA GH9 module [Caldicellulosiruptor bescii] |
| 1JS4_A | 7.76e-51 | 348 | 768 | 4 | 440 | EndoEXOCELLULASE:CELLOBIOSEFROM THERMOMONOSPORA [Thermobifida fusca],1JS4_B EndoEXOCELLULASE:CELLOBIOSE FROM THERMOMONOSPORA [Thermobifida fusca],1TF4_A EndoEXOCELLULASE FROM THERMOMONOSPORA [Thermobifida fusca],1TF4_B EndoEXOCELLULASE FROM THERMOMONOSPORA [Thermobifida fusca],3TF4_A EndoEXOCELLULASE:CELLOTRIOSE FROM THERMOMONOSPORA [Thermobifida fusca],3TF4_B EndoEXOCELLULASE:CELLOTRIOSE FROM THERMOMONOSPORA [Thermobifida fusca],4TF4_A EndoEXOCELLULASE:CELLOPENTAOSE FROM THERMOMONOSPORA [Thermobifida fusca],4TF4_B EndoEXOCELLULASE:CELLOPENTAOSE FROM THERMOMONOSPORA [Thermobifida fusca] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02934 | 1.04e-50 | 348 | 792 | 76 | 534 | Endoglucanase 1 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celI PE=1 SV=2 |
| P26224 | 3.98e-50 | 347 | 769 | 29 | 464 | Endoglucanase F OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celF PE=3 SV=1 |
| P26221 | 9.56e-50 | 339 | 768 | 41 | 486 | Endoglucanase E-4 OS=Thermobifida fusca OX=2021 GN=celD PE=1 SV=2 |
| P22534 | 3.22e-48 | 348 | 784 | 26 | 475 | Endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celA PE=3 SV=2 |
| P26225 | 1.29e-47 | 348 | 768 | 37 | 474 | Endoglucanase B OS=Cellulomonas fimi OX=1708 GN=cenB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
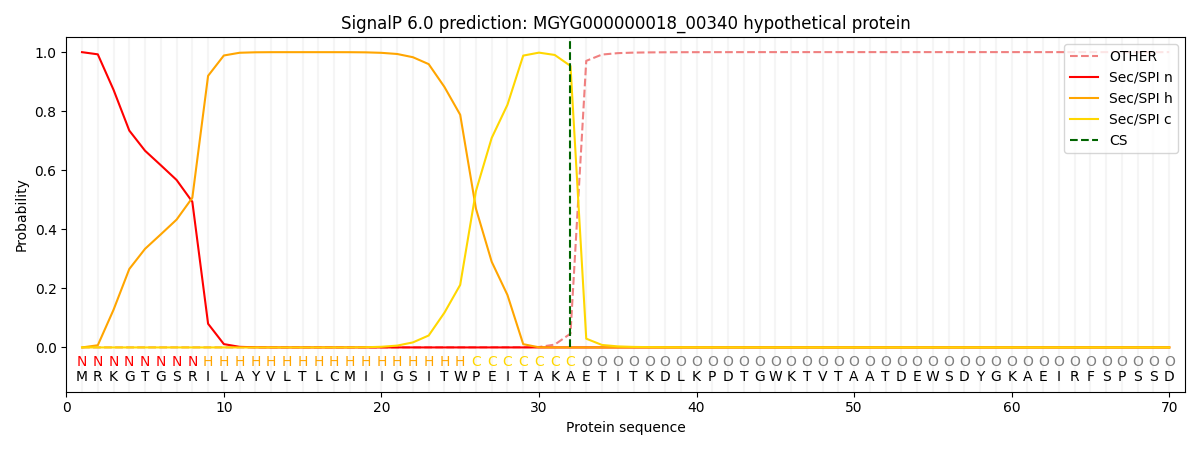
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000867 | 0.998215 | 0.000292 | 0.000263 | 0.000181 | 0.000162 |