You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000059_01577
You are here: Home > Sequence: MGYG000000059_01577
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lawsonibacter sp900066645 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; Lawsonibacter; Lawsonibacter sp900066645 | |||||||||||
| CAZyme ID | MGYG000000059_01577 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 398097; End: 405269 Strand: - | |||||||||||
Full Sequence Download help
| MSHTPNVPMD DAHLTKYAVN AARELKPRGK VPVRRLVRRL KRDLSILNRI QEELSEWSQG | 60 |
| KAVLPPSVEW LLDNHYLAVR EGEEALRALK KAGPLRGDGQ GGALLQRCVR GGVWAVPHLE | 120 |
| RERLTWYLKA FQTVQPLTER ELSLLVPVLA IELIQELAQE AGQLEELRQG RTDPGRLEHL | 180 |
| FSALRALEGE NWGPLLEEIS RVEEILTQDP SGHYPHMDDD TRRRYRQQVC RLAKKFRLEE | 240 |
| SQTARKALEL AKRGTGAARH IGWYLYRQPL GREKRLPSGL GYGGAVVGLS LALAAGLWVA | 300 |
| AGSLWLSLLL LLPLSDIVKN VLDFGLVRIV PPRPVPRMEL EGGIPRENRT LCVVVSLLTG | 360 |
| EDSGPKLAAL LERYRLANRD SGPELRFGIL ADLPDGAAPM GEEGAAWVTS AHRAIQALNE | 420 |
| KYGGGFYLFF RRPAFSAQDE KYMGWERKRG ALTELVRLLK RKPTGLRTEV GEEDWLREVT | 480 |
| YVITLDGDTS LNVGTAREMV GAMAHPLNQP VVDGQRRVVT SGHALFQPRV AVELEAANRS | 540 |
| FFSRVYGGLG GVDPYGSTAS DVYHDLFDQG TYTGKGIFQV EAFFTCLDGR FPENAILSHD | 600 |
| LLEGSYLRAG LLGEVELTDG CPYKVYGYFA RLHRWIRGDW QLLPWLKNQV PDGQGGTEDN | 660 |
| PLSLLARWKI FDNLRRSLSP VGTLAALVLG ICFSGRAFAW AGGVAVLAAA SNLLLSGAEL | 720 |
| AARRRNGKHR RYHSTVIAGL AGVILQTALQ LVLLPYQAYT AATAAATALW RMGVTKRGLL | 780 |
| AWVTAAQAEK GRDGLWFYGK KCWFSVVIGG AVLLLAQLRI GRLVGVVWLM APALAWAISR | 840 |
| PGKKGKELPP TDRSFLLHQG ALIWGYFDAF LRQEDHFLPP DNWQEQPSQG LARRTSPTNI | 900 |
| GMALLSVMAA ADLKLITRER GIDLISHILD TIEELEKWRG HLYNWYDTAS LRPLQPRYVS | 960 |
| TVDSGNLRGC LIALREGLYQ WGEDVLARRA EHLSDAMDCA PLYDRERRLF SIGYEAERDR | 1020 |
| LTPGWYDLMA SEARQTSFIS VARGEVSPRH WRRLGRMMLG DNDYSGMASW TGTMFEYFMP | 1080 |
| NLLLPCEPNS FMYESLSFCV YAQKRRGNKT HTPWGISESA FYAFDPGMNY QYKAHGVQAL | 1140 |
| GLKRGLDSEL VVSPYSSFLA LLLAPRSALR NLRRLRDMGL EGPYGLYEAV DYTPARMTEG | 1200 |
| QDHEVVRSYM SHHLGMSLIA IDNALNDNVM QRRFMKDCDM AAYRELLQER VPVGAPIMRQ | 1260 |
| TERDIPEKLR PVQGPALVRA GREFGRLAPE CHLVSNGSWS VLACDNGLTA SRMGDCQITL | 1320 |
| AKLGQYYAPA GISLFFQGRE GIFGLTPAPL YQKGEYSWEF HSAGAAWTFT WEGLATRTTL | 1380 |
| AVPRRENGEL RRVELSWTGE GRLEGELLAY LEPVLCPLAD FQSHPAFSRL FLEGELDDHG | 1440 |
| VLFQRRPRGE EKSLSLAVLW EGEGITACLD RASALGRGGL RALGGRRAGP LERGVGTDPC | 1500 |
| LMVRFPVLLE PGERAVFALA LGAGDTPAAA STGARNILRG QAGEAASLAP LARKLSLTER | 1560 |
| QGIEAFQLLS RLSAAEEPGE RPPKSALWPY GISGDLPLAV GPVTEGEGRE QAVLWCRWHQ | 1620 |
| LLTRSGYPFD LVLLVEEGGD YRRPLRTALT EELKTMGAES ALGAKGGIHL ADSGAAPAAL | 1680 |
| AWARVILPMG PGEQREEKLS PATPVRLSLA QPEWRAGEEG TVLIRTGKAL PPVGWSQVLC | 1740 |
| NSQFGWLTDE TGNGLLWTGG NSREGKLTPW SNDPLAIGGP EKIMVSLEEK EWSVFADGDG | 1800 |
| CPCTVTYGPG FARWEKQMGT GRLVTEAFVP LIGTKRILTF TLAGTSGTLS YRMGEGEPMA | 1860 |
| RPLAQGLPLT LVTEPGHRGL ESRFSTKRYL DLRGETLTWW RERVSSLTLS TPDRTLNHYL | 1920 |
| NGWCLYQVTA CRLMARTSQY QNGGAFGFRD QLQDVAALLY TWPQRTREQL LLAASRQFEE | 1980 |
| GDVQHWWHPP AGAGVRTRIS DDLLWLPWVL CRYCSVTGDW EVLKEQVPYL TSRPLEPKEM | 2040 |
| ERYEVPQVSS KTDTLYHHAL SAIQCVLDRG TGSHGLARMG GGDWNDGMNR VGAAGRGESV | 2100 |
| WLTWFLTMVL QDFAPLCQRS GEEELAQELL AQGEKYLAAA QAAWDGNWYR RGYYDDGTPL | 2160 |
| GSRESDQCQI DSIAQSFSVL PQGADRERAD RAVSAALTRL FDRERQVVRL FHPPFDGKGR | 2220 |
| KDPGYIKGYP AGVRENGGQY THGAVWLAMA CFRLGRAGEG WEVLRALLPA CHATEGYRAE | 2280 |
| PYVLAADVSY ATGREGQGGW SWYTGAAGWY WQVAVQELLG LKVKEGRLRV QPRLPFDWPG | 2340 |
| YQAEWRLERG TLLIRVERGE ADAALLDGQP VDEVALKELE GEHHLRVVIP | 2390 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH94 | 1897 | 2384 | 5.7e-184 | 0.46525096525096526 |
| GT84 | 1025 | 1237 | 3.2e-87 | 0.986046511627907 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3459 | COG3459 | 1.84e-160 | 1900 | 2389 | 565 | 1052 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
| pfam17167 | Glyco_hydro_36 | 1.17e-103 | 1899 | 2322 | 1 | 425 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
| cd11753 | GH94N_ChvB_NdvB_2_like | 1.96e-32 | 1230 | 1530 | 1 | 317 | Second GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This second of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam06165 | Glyco_transf_36 | 2.08e-21 | 1288 | 1522 | 2 | 247 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
| pfam10091 | Glycoamylase | 4.30e-16 | 1024 | 1236 | 1 | 213 | Putative glucoamylase. The structure of UniProt:Q5LIB7 has an alpha/alpha toroid fold and is similar structurally to a number of glucoamylases. Most of these structural homologs are glucoamylases, involved in breaking down complex sugars (e.g. starch). The biologically relevant state is likely to be monomeric. The putative active site is located at the centre of the toroid with a well defined large cavity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QBB66089.1 | 0.0 | 1 | 2385 | 1 | 2395 |
| ALP93881.1 | 0.0 | 1 | 2080 | 1 | 2094 |
| BCI59942.1 | 0.0 | 47 | 2358 | 40 | 2444 |
| AVX19642.1 | 0.0 | 34 | 2389 | 34 | 2765 |
| AVX30049.1 | 0.0 | 34 | 2389 | 34 | 2765 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3QDE_A | 8.02e-50 | 1883 | 2357 | 290 | 771 | ChainA, Cellobiose phosphorylase [Acetivibrio thermocellus],3QDE_B Chain B, Cellobiose phosphorylase [Acetivibrio thermocellus] |
| 2CQS_A | 7.85e-48 | 1824 | 2372 | 259 | 823 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQS_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQT_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],2CQT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],3QFY_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFY_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFZ_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QFZ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus] |
| 3ACT_A | 5.76e-47 | 1824 | 2372 | 259 | 823 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Histidine mutant [Cellulomonas gilvus ATCC 13127],3ACT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Histidine mutant [Cellulomonas gilvus ATCC 13127] |
| 3AFJ_A | 5.76e-47 | 1824 | 2372 | 259 | 823 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127],3AFJ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127] |
| 3ACS_A | 1.35e-46 | 1824 | 2372 | 259 | 823 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase W488F mutant [Cellulomonas gilvus ATCC 13127],3ACS_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase W488F mutant [Cellulomonas gilvus ATCC 13127] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 0.0 | 59 | 2374 | 81 | 2818 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
| B9K7M6 | 1.42e-49 | 1895 | 2357 | 304 | 773 | Cellobiose phosphorylase OS=Thermotoga neapolitana (strain ATCC 49049 / DSM 4359 / NBRC 107923 / NS-E) OX=309803 GN=cbpA PE=1 SV=1 |
| Q76IQ9 | 3.83e-37 | 1900 | 2389 | 297 | 799 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio proteolyticus OX=671 GN=chbP PE=1 SV=1 |
| Q7S0S2 | 2.44e-35 | 1875 | 2390 | 281 | 791 | Cellobionic acid phosphorylase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=NCU09425 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000069 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
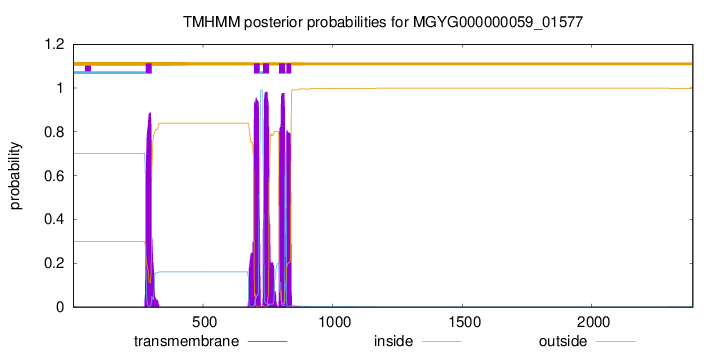
| start | end |
|---|---|
| 280 | 302 |
| 697 | 719 |
| 732 | 754 |
| 794 | 816 |
| 823 | 840 |
