You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000131_00856
You are here: Home > Sequence: MGYG000000131_00856
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Anaerotruncus rubiinfantis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Anaerotruncus; Anaerotruncus rubiinfantis | |||||||||||
| CAZyme ID | MGYG000000131_00856 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 400393; End: 407715 Strand: + | |||||||||||
Full Sequence Download help
| MKEIEKVTIT EKVEGLFSQL SEHRNTVAGM RRELCKNLDV LRESFAALRE TGAAAGWQQW | 60 |
| LSDNFYALEG HAKQSMNDLR GYRKNSVPLA SLYFILRGVF ADAVVPLTQE NTAEALGAAN | 120 |
| RLQELGERQF GFVYPALKCT LIAAAAMACS PNLADEERER RISFAVVGLS QLYEIDFAEL | 180 |
| TEQCSTAEEI LLEDPSGVYP KMARQSKRYY REIAARIAKK SGAVESAVAR DVVEYAKKSD | 240 |
| GERTSHVGYY LLEKDPQAKK GKLHGAAALA ATWIVPVFLC ALCGFCWDRI FLPVLLYLPL | 300 |
| VEIVRIFACD LAMRGIPARH IPRMRLGGEA PRTVVVISTL LPAPEKARDL AKRLEQLYFS | 360 |
| NKGEHLRFCV LADFKEDRRP YHPQDAVQIG AAKKVVEELN ACYGNRFMLF VRRRSFCPTQ | 420 |
| DRYCGWERKR GAITEFVRFL NGSETSVSCF VGDRAALSEA RFLIALDSDT NLLFDTADVL | 480 |
| ISAAMHPLNR PVVGKNQVVT AGYGIFVPAI GTELDSAKET DFTRIMAGAG GISTYEQESN | 540 |
| DFYQDLFGEA IFSGKGLIDI EAFRTVVGHR FPENALLSHD ILEGAYLRTA LISDVEMTDG | 600 |
| MPASALSWFS RLHRWIRGDW QNLRFIGKRF SVNGIKRVNP IGGLSRYKML DNLRRSLTPV | 660 |
| AAVLLVAAAG LARPDEALWL CAAAFLSVSL APLLAALRAF LAGGRFALTR RYFARTLPRV | 720 |
| FELAGQALLS FVMAAQQAVV TLDAVFRTLW RMLVSRKNLL QWTTAAQGEL REVSFSDAAR | 780 |
| RSWISELLGL LLLIFAPRTH LIVKLCGGLF LLYAPLAVMT ARRTVERHPV PDPVQRERIL | 840 |
| SWCAQMFRFY EDYANEENHF LPPDNVQFSP KKTVARRTSP TNIGMMLLSL LAARDFHLLD | 900 |
| SRELYLRLDR TFETVESLET FHGNLYNWYA TDDLRVLPDG FISAVDSGNY LCALVALRQG | 960 |
| VLEYAPQEPL LREIAQRIEK ILQGTDLSIF YNRRRRLFSI GVDQNGNQVG SHYDFLMSEA | 1020 |
| RTLSYYAIAT RQASRKHWRA LNRTMSRSGV YAGPVSWTGT MFEYFMPHLL LPVYEGSLLG | 1080 |
| EAMQYAIYCQ KKRARQAGVP WGISESGYFA FDEALNYQYK AHGVQKLGVK QGLDRELVVA | 1140 |
| PYASFLTLPF DPDASMQNLE RLERMGITGQ YGFYEAADFT PVRVPQGERY AIVRSYMAHH | 1200 |
| VGMSMIACAN ALLGNVMQKR FMRDHAMAAA RDFLQEKIAK DAVVYDQMKP ENNERTKSAP | 1260 |
| KENETISPPV SALSPKAALL ANGVMSHAFS STGVSFLRYG DNDVTRRSRD ALLHGQGILS | 1320 |
| VLRMNGETVA ASEAPFYQSG VERHCTFGPE FVSFSAKAGT AELEQRFTLD GILPVSRCDA | 1380 |
| TVRNHTGNKA VAELLFYFEP VLSRESEYAA HPAFSKLFVL GERDAATDTI HFLRRHRDCT | 1440 |
| DGLHLSVGFE QTVEYNFSLR REEVTPYPDG LAHLLRFDTA PLTGKTATPD GCCALRVTLV | 1500 |
| VPAHGEETVG LLLSCAPTQA ESIGNLLTIR DGGRTAAQSL FLNRSITGRI GMEVLPDLLF | 1560 |
| GAEADETQQK ALAENSRGQD ALWSLGISGD LPIVVFDWEK KPDEACLLAY AEFWQAMKLH | 1620 |
| RLDFDLCVLG RPELALPQGV RKIDPAAYEP ELVTALRAAA CHTAGAERNA GTIKGFSPMA | 1680 |
| VLHTSPLPIP QEEGRFDLVG GAYVKGRFYV ERVTPLPFSH ILANRNFGTL LHDCGLGNTW | 1740 |
| WQNARECRLT PWQNDIATGN DGERMLLRIG EEIYDLCCGA RASFSPAGAW YDGMAGGIRT | 1800 |
| KLQVQVAETE SLKIMDVTLQ NDTDEEVEII CAYVIEPVLG VSRLTAKYTQ FTQENGCLTL | 1860 |
| HNPYTMELPA YAAVCTPGEH PSYTVDRTAF WSGDWSAREL LPNNDPIAGT VVRKRLPPRR | 1920 |
| REKIRFILAA AAKRDAAVKL ATGVLPPSGG VRPGISIATP DLALNHFVNG FLPHQVVAGR | 1980 |
| LFGRCAFYQC SGAYGFRDQL QDAAACLLID PELTKEQILR CCAAQFPEGD VLHWWHDLPG | 2040 |
| GVRGVRTTFS DDLVWLPLVV CDYLKATGDE SILSIPVAYC EGEPLGPGEQ ERYQRVHASA | 2100 |
| MKEEVFFHCV RAIEHACNLG DKGIPKIGCG DWNDGFSNIG TQGKGQSVWL GMFLAIVFDR | 2160 |
| FAPILEARGD SYRASVFRGN AENLRGSIDQ NCWDGGWYLR AFFDDGTPVG SRENSECRID | 2220 |
| LLAQSFSVFC GMPDNARVME SLDAARKRLV DDRGQIIKLF TPPFDQSTNN PGYIKAYPTG | 2280 |
| IRENGGQYTH GAIWFAMAML EAGRTDEGYR LLSYLNPAGR CTDAGLAQAF KTEPYYMPAD | 2340 |
| IYTHRNCYGH GGWSIYTGAA GWYYRAVVET LLGIRFCGDH LEVFPRLPEG WTRFSAQVRR | 2400 |
| SGYEIELEVM KTGVESITVD GTPAGKIPLD GKNHNVLLTF | 2440 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH94 | 1559 | 2436 | 3.9e-213 | 0.8841698841698842 |
| GT84 | 1011 | 1224 | 8.6e-85 | 0.9906976744186047 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3459 | COG3459 | 7.55e-156 | 1556 | 2440 | 116 | 1054 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
| pfam17167 | Glyco_hydro_36 | 1.29e-96 | 1955 | 2375 | 9 | 425 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
| cd11756 | GH94N_ChvB_NdvB_1_like | 4.32e-33 | 1694 | 1890 | 9 | 217 | First GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This first of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam06165 | Glyco_transf_36 | 8.59e-25 | 1715 | 1928 | 1 | 246 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
| cd11753 | GH94N_ChvB_NdvB_2_like | 1.52e-24 | 1217 | 1509 | 1 | 304 | Second GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This second of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCO21007.1 | 0.0 | 411 | 2439 | 1 | 2058 |
| BCI59942.1 | 0.0 | 11 | 2417 | 5 | 2450 |
| AVQ95160.1 | 0.0 | 1 | 2438 | 11 | 2549 |
| AYF37850.1 | 0.0 | 1 | 2438 | 11 | 2549 |
| QCN91406.1 | 0.0 | 1 | 2438 | 11 | 2549 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ZLE_A | 1.25e-47 | 1906 | 2425 | 250 | 775 | Cellobionicacid phosphorylase - ligand free structure [Saccharophagus degradans 2-40],4ZLF_A Cellobionic acid phosphorylase - cellobionic acid complex [Saccharophagus degradans 2-40],4ZLG_A Cellobionic acid phosphorylase - gluconic acid complex [Saccharophagus degradans 2-40],4ZLI_A Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex [Saccharophagus degradans 2-40] |
| 3QDE_A | 2.47e-45 | 1714 | 2436 | 20 | 806 | ChainA, Cellobiose phosphorylase [Acetivibrio thermocellus],3QDE_B Chain B, Cellobiose phosphorylase [Acetivibrio thermocellus] |
| 3S4C_A | 8.16e-44 | 1714 | 2423 | 20 | 801 | Lactosephosphorylase in complex with sulfate [Cellulomonas uda],3S4D_A Lactose phosphorylase in a ternary complex with cellobiose and sulfate [Cellulomonas uda] |
| 3RRS_A | 1.91e-43 | 1714 | 2423 | 20 | 801 | Crystalstructure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RRS_B Crystal structure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RSY_A Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3RSY_B Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3S4A_A Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4A_B Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4B_A Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda],3S4B_B Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda] |
| 2CQS_A | 2.00e-41 | 1956 | 2423 | 343 | 821 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQS_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQT_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],2CQT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],3QFY_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFY_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFZ_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QFZ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 2.42e-300 | 162 | 2422 | 285 | 2821 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
| B9K7M6 | 1.01e-43 | 1694 | 2436 | 4 | 808 | Cellobiose phosphorylase OS=Thermotoga neapolitana (strain ATCC 49049 / DSM 4359 / NBRC 107923 / NS-E) OX=309803 GN=cbpA PE=1 SV=1 |
| Q7S0S2 | 1.06e-41 | 1955 | 2440 | 319 | 790 | Cellobionic acid phosphorylase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=NCU09425 PE=1 SV=1 |
| Q8P3J4 | 8.15e-41 | 1930 | 2421 | 308 | 773 | Cellobionic acid phosphorylase OS=Xanthomonas campestris pv. campestris (strain ATCC 33913 / DSM 3586 / NCPPB 528 / LMG 568 / P 25) OX=190485 GN=XCC4077 PE=1 SV=1 |
| Q76IQ9 | 1.95e-40 | 1957 | 2440 | 306 | 800 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio proteolyticus OX=671 GN=chbP PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
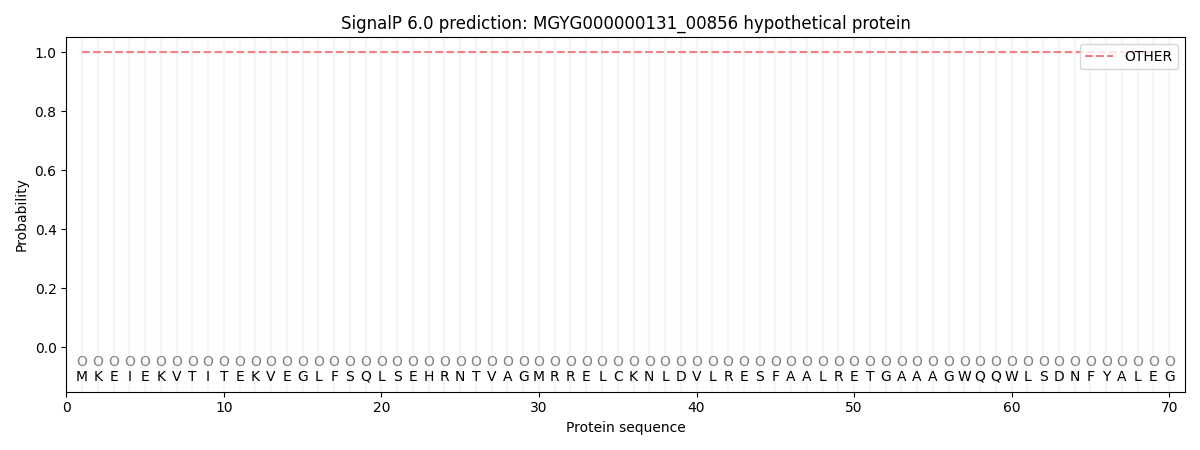
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000047 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
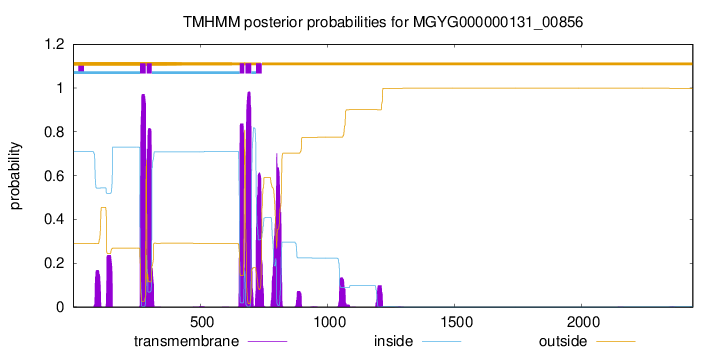
| start | end |
|---|---|
| 263 | 285 |
| 290 | 309 |
| 656 | 673 |
| 678 | 700 |
| 720 | 742 |
