You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000898_00492
You are here: Home > Sequence: MGYG000000898_00492
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
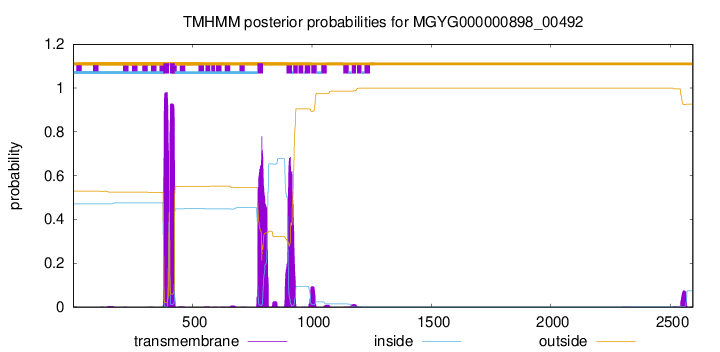
TMHMM annotations
Basic Information help
| Species | UBA5394 sp900542615 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-138; UBA5394; UBA5394 sp900542615 | |||||||||||
| CAZyme ID | MGYG000000898_00492 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 387; End: 8174 Strand: + | |||||||||||
Full Sequence Download help
| MAQNRTYFGL QDVTLEGEEL CREAAQRAAW STSRRRLRWG TKNVLSIGED RRTIMAAYKQ | 60 |
| ARERAAQGLE LPQAMECLLD DLYVAEKALT SLRDEDSGQW RGLPCMAQGE RLGLPRVYDM | 120 |
| AVCLIGHRAG RLEESMLTRF LEAFQGVAPL KLAELCALPA MLRVALVKLI ALECEGALDA | 180 |
| MGQYAQAEAM AAQAGRGNPQ REREAYLHKP YLAARLFGLL SEAGEQGACS RMLRRLEQAD | 240 |
| LDLEALCANL QREDGIRGDR LRHAIKSLRT LDAMDWEKSC EGFSLVDRAL RGDGAYPGMD | 300 |
| ARSRAWYRQR VEALAARLGV AETVVARQAV SLGQARQEQG GRAAQAGYYL LEEGQRDLYA | 360 |
| ILRPDKRCRL QSEERKLWRF WIVQGALLAG LVYLCGAAAW YRGLLALLPA WSLAAGLSVR | 420 |
| LFLARSQPSM LPRMEYKDAL PPEQATLVVV PALITDAESL RGVIRQLEVH MLATRMEHCY | 480 |
| YGVLGDFPDG KQAQRPGEGE LLRLAKSLVE ELNQKYPSPT PLFYYLHRRR TYQKADGLYM | 540 |
| GWERKRGALC QLVTLLTQGD ATPFALLTHP LPQGIRYCLT LDADTVLPPG ALAKLVGAMA | 600 |
| HPLHAPEWDD QGVVKKGYGI LAPRMAALPR GAAKSGFAWV ASGDSGLDSY FPLCGEFYQD | 660 |
| VFGAGIFGGK GIFHVEAFAR ALPRWIPENQ VLSHDLLEGC FLRAGLVEDV TLYDCEPAGF | 720 |
| IAWWKRQHRW TRGDWQLLPY LGDGVRDGAG ILRDNPLSSL SRYKIFDNLR RSLTPLGALA | 780 |
| CMLALPYLGW GWYGALALLC ILEGPVLEAV CLPVQLLRSR RPVRLGGALL DRMPGAARAA | 840 |
| LDLLVLPYGT ARLADAQART LYRVVHSHKH MLQWQTAAQT RGKPGSLGAY YSAMWPQTAI | 900 |
| GLLMLAGMAL GHAPWISGLL GALWLAAPWG IERLDRGKAP QPLSPRAREL LLDIARRTWA | 960 |
| FFDGFAGPQT GWLPPDNYQQ APARPVVANT SPTNIGMGMI ACVCAMDLGF LTGEQLCRRI | 1020 |
| GNMLDTLEGL ETWRGHFYNW YSLWDRRVLS PRYVSTVDSG NLAACLLTVA AALEELPGDM | 1080 |
| GRAQGERCRA MAKAMDFRAL YDQERGLFHI GFDEGAGRLS RAWYDLMASE SRLTSLVAVA | 1140 |
| LGQVEAEHWF HLGRLLVPAG GGRALLSWSG TMFEYLMPVL FTGLVPGTLL HESCVNALQA | 1200 |
| QIRYGQAQGF PWGVSESGYY AFDRAMYYQY RAFGVPRLGL MAQREPSRVV SPYSTLLALG | 1260 |
| VPGWEEKALA NLERLVDEGA LGEYGMLEAL DYTGSRVAAG KERELVQSYM AHHQGMGLCA | 1320 |
| LTNCLCGDSI RRRFMALPQI RAVEILLEEK PPAHGIVIRE FESAALQGKA PEKKPHRPRR | 1380 |
| VTGPKAVPET QLLSNGEYTL FLADSGLGFS KWQDVLLTRW RPDPLRGDGG IHLMARLGEE | 1440 |
| TWELTWGAET ILHPHKVEFH GRRGPLSTHV ETYVCPQLDG EIREITLGNH GEEKLEVELG | 1500 |
| VFGEVCLATA GEDMAHPAFV RLTVEAAQRE GMLLFWRRQG GGAKPKGVLY AQLYAPGLRP | 1560 |
| RYCTDRFSAR GRGATLGESM RRPLGGGPAE APVDPCIAAR ATVALEAGQS RTLWFLMGYA | 1620 |
| QSEEKALAQA EDLRAGLGEW EELAWAHALS DLRMAGLSEG KAELFQRIAA RLLLQIPQKP | 1680 |
| QRPKDAPLGP GLEGLWQLGV SGDLPILLME VESLQGLRMA RTLLEFCSYM AAQNCPIDLV | 1740 |
| LVGCYPHAYR GELQLRLGEL CSRHPQAKLL HSYALTQEQR QLLRDMALVV ADGRPGRSLD | 1800 |
| KQFAQEEAPS WPPQVLTPGL EPMGEAALPP LPPLAFANGF GGLDTASGAY VIQLGPGEKT | 1860 |
| PMPWCNILTN GRFGALVSES GGGYTFGENS RTDKLTPWQN EPLSDRRGEI LLLWDGQEGH | 1920 |
| RPFTIEPGRL QREPARVRHG YGYTAFSTTA YGLNCEAVTF IDPTAPVKYT LLTLENPGKQ | 1980 |
| ERRLQLLYQA EWALGERGDP ASIYAYTHGE AAFARSLRTP AEAPGYLACP TLPVQVCHDR | 2040 |
| EALLKGGWWA EALPEAPRHY GGAMSGLRGG MQIPAGGRAQ VVLMLGQEAE AEAAESISRS | 2100 |
| TPSYVAERLQ RVEADWQARL GKIRVQTPDP AFDALINGRL LYQVYAARLM ARTGYYQCSG | 2160 |
| AMGFRDQLQD MLALLQVEPA RVREQLLLCA GRQFPAGDVL HWWHMPMRGV RTRIVDDRLF | 2220 |
| LPYVLSYYLE TTQDMDILDV PVAYLADRPL AEGQRDLYDR MEPGEAAEPL YGHCMRAIRS | 2280 |
| ASRFGAHGLP YMEGGDWNDG MDQVGQGGGE SVWLGWFLLA VYRRFAPIAR AYGRDADAEE | 2340 |
| LEGAIPGLQA ALEAAWDGAW YRRAYFADGT PLGSASNHSC RIDCISQAWA AICGGAHGAE | 2400 |
| AMDALMEMLL DPQSGILRLL TPAFRESLEQ GNPVGYITAY VPGVRENGGQ YTHGAAWAVW | 2460 |
| ACCCLGRSEE AFRLFETLNP LVHTSTRTGA VRYGVEPYAL AGDVYAPPNG GRGGWSWYTG | 2520 |
| AAAWLYKIGL EDMLGIRRQG QELCFAPCVP FDSFAVTYAF GSATYILRFQ RGEKKGPKRI | 2580 |
| ALLDDGKCHT VKVVF | 2595 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH94 | 1642 | 2582 | 3.8e-234 | 0.8918918918918919 |
| GT84 | 1123 | 1338 | 1.2e-82 | 0.9906976744186047 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3459 | COG3459 | 0.0 | 1562 | 2587 | 2 | 1031 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
| pfam17167 | Glyco_hydro_36 | 1.57e-103 | 2116 | 2537 | 2 | 425 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
| cd11753 | GH94N_ChvB_NdvB_2_like | 1.16e-65 | 1330 | 1631 | 1 | 322 | Second GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This second of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| cd11756 | GH94N_ChvB_NdvB_1_like | 8.11e-58 | 1836 | 2087 | 4 | 269 | First GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This first of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam06165 | Glyco_transf_36 | 1.59e-37 | 1387 | 1618 | 2 | 247 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVX19642.1 | 0.0 | 15 | 2595 | 8 | 2766 |
| AVX30049.1 | 0.0 | 15 | 2595 | 8 | 2766 |
| QSQ07709.1 | 0.0 | 18 | 2595 | 5 | 2856 |
| APV50312.1 | 0.0 | 47 | 2586 | 65 | 2815 |
| ATW26012.1 | 0.0 | 11 | 2595 | 32 | 2712 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2CQS_A | 3.81e-49 | 1840 | 2586 | 23 | 819 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQS_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQT_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],2CQT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],3QFY_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFY_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFZ_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QFZ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus] |
| 3ACT_A | 2.81e-48 | 1840 | 2586 | 23 | 819 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Histidine mutant [Cellulomonas gilvus ATCC 13127],3ACT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Histidine mutant [Cellulomonas gilvus ATCC 13127] |
| 3AFJ_A | 3.74e-48 | 1840 | 2586 | 23 | 819 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127],3AFJ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127] |
| 3ACS_A | 6.61e-48 | 1840 | 2586 | 23 | 819 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase W488F mutant [Cellulomonas gilvus ATCC 13127],3ACS_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase W488F mutant [Cellulomonas gilvus ATCC 13127] |
| 3RRS_A | 2.26e-46 | 1840 | 2584 | 3 | 798 | Crystalstructure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RRS_B Crystal structure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RSY_A Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3RSY_B Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3S4A_A Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4A_B Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4B_A Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda],3S4B_B Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 0.0 | 49 | 2582 | 64 | 2818 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
| B9K7M6 | 1.57e-52 | 1840 | 2595 | 3 | 812 | Cellobiose phosphorylase OS=Thermotoga neapolitana (strain ATCC 49049 / DSM 4359 / NBRC 107923 / NS-E) OX=309803 GN=cbpA PE=1 SV=1 |
| Q8P3J4 | 6.95e-32 | 2117 | 2586 | 319 | 773 | Cellobionic acid phosphorylase OS=Xanthomonas campestris pv. campestris (strain ATCC 33913 / DSM 3586 / NCPPB 528 / LMG 568 / P 25) OX=190485 GN=XCC4077 PE=1 SV=1 |
| Q7S0S2 | 1.27e-23 | 2107 | 2575 | 299 | 759 | Cellobionic acid phosphorylase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=NCU09425 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
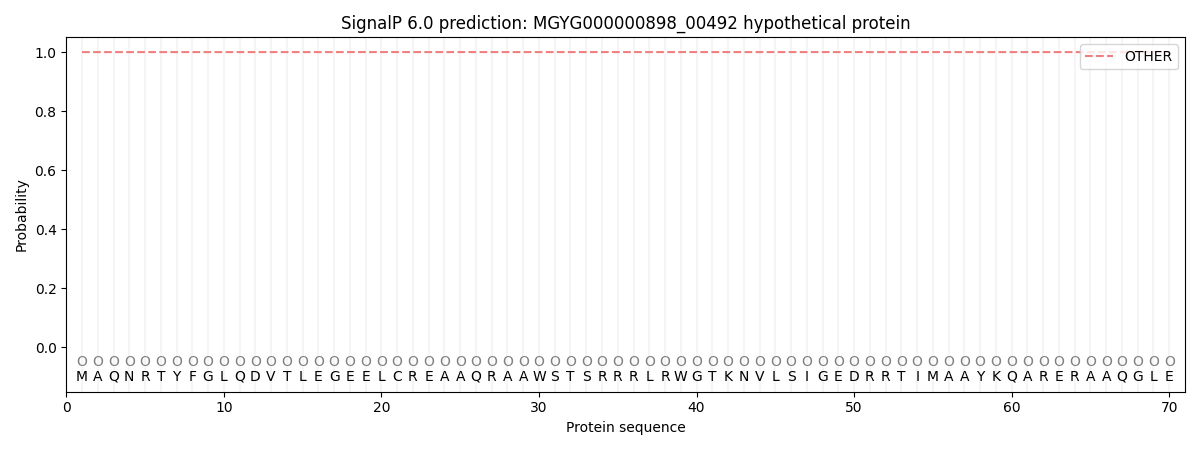
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000066 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |