You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001043_00264
You are here: Home > Sequence: MGYG000001043_00264
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-877 sp900753325 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RF39; UBA660; CAG-877; CAG-877 sp900753325 | |||||||||||
| CAZyme ID | MGYG000001043_00264 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 85446; End: 93086 Strand: - | |||||||||||
Full Sequence Download help
| MRDALSKFFK DFKKINDYYN YLINKTKNAC YVGITNEWII DNFYLLAEHK TNISNDKKHI | 60 |
| TKELSHTNSI YYCLKQIAIS NNYSVSFKTL VSSLKAYQKE NNTNFSYREI SAIKYLLLFI | 120 |
| YTEKLVLLCN DEKNKLIIQE KIAKIIDNCD KDNVKLDEFI NNNFDVSNNQ YYIYELNQQL | 180 |
| RKLGSRSNSL FKEINELLRT KNISLKEIIN DFHQQKIDND LLVSNIFNDI KALFELSDEE | 240 |
| LYEKVSVVEK ILLEDDIYKK MTVESKDIYR KQIRKLTRIK RTTELSLVNG LFKKSREENK | 300 |
| HIGFYLFRNK NYQFRTIMYL LVITLLTFII SFFLSKYFIN MRWLGFLILL VPINQLVIKV | 360 |
| INFLLTGLIP ATVLPKIDYS KGLSKDATTM VVIPTIISDT KKVKEMFDVL ETFYLINKSD | 420 |
| NLYFTLLGDA KASDKKDEKC DKEISLYGEE LANSLNKKYG RSLFYFIYRK RFFNESENCY | 480 |
| LGYERKRGAL LQFNKVLLGK MSDSASKKNF HINTLEKTNL DIKYVITLDT DTRLVLSTAL | 540 |
| NLVGAMAHPL NKPVLNKDRT KVISGYGIMQ PRVGVDIEAT NKSLYSQIFA GVGGFDTYSA | 600 |
| VASNVYQDFL GEGSFVGKGI YDLKVFDEVL ENTFPDNLIL SHDLLEGNYL RVAYVSDVEL | 660 |
| IDDFPSEFLT DTTRHHRWAR GDTQIIGWLF NTVRNKNNKK VHNPINLLGK FKILDNIVRM | 720 |
| FLMPSLLLIL ILSVFTNYYL WWIGFVVLEI AISIIFFLSS KVSLKNNKKM TVYYKNLLAG | 780 |
| GRSLLYRAFI TFSDIPFYSR LYLDAFFRTL YRLFVSHKNL LNWVTAEDVQ KNAHSDLLTY | 840 |
| IKNFIPNFVC SLIFIILGIF SKSYVYYIIA FIFLIAPVIL YRVSLTINHG ALEISEEKCE | 900 |
| KLRKLTFDTW MYFSDNLKEE YNYLIPDNYQ ENREIKLDMR TSPTAIGYSL TSVVCAYEMK | 960 |
| FIDEDRALFL IKNILESVDS LEKWHGHLYN WYDIKSMEVI NPRFVSTVDS GNFIAGVIVV | 1020 |
| KEFLKKFDQD KLIRLCDKLI NNANFKKLYT KRCVFSIGYD DNEGRLSIYN YNKFASEARL | 1080 |
| TSYIAICKGD VSAKHWFCLD KSLTTYKGRK GLISWSGTSF EYYMPLLFMK NYPNTLLDES | 1140 |
| YYFAYFCQKE YIEKVSRKLP WGISEAAYNE LDNSLNYKYM AFSTPYLKAK DDKNNRIVLA | 1200 |
| PYASIMALEL FPDEVVSNLE KFKNLNMLGK YGFFESYDYD NKGVVQAYFA HHQGMILIGL | 1260 |
| VNYLKAFAIK NYFHNNVSIR TFDILLKEKV QLKTDIDMKM ASYKRYNYNK EKVENDIRTF | 1320 |
| NYISYMPEVS VLSNKKYTLL MNDRGNSFSR YRTLQLNRYR KVTEQDYGIF MYIKDLKTNY | 1380 |
| VWSNTFAPVN KSSDNYEVVF ASDKIKYLRV DGSISTATEI VVCPNHHAEI RKITFKNNSD | 1440 |
| SPRMLELTTY TEVILSENMD DVSHRAFNNM FVSSEFDNKN NALIMKRKNR GDSNVNNYMV | 1500 |
| SRLIINNPLD KYSYETERSK FIGRGNTVSN PISLNSKLSN YTGDNLDPVS SIRNRIVVEA | 1560 |
| NSSKEVYLLV GFGRSREQIN DIIKSYSDSK EINKVFRVAA LANAFNMKSM NITGDDLRLF | 1620 |
| NIMLNYLYQT TKISVSEERM DLLRKNSLSQ TGLWKFGISG DRPIILVDIS DISDLPFVFS | 1680 |
| ILKAFEYYKN NSIFVDIVIV NNENEQYSEV INREIDDEIY RMYSVNSFYH TPGIIKVIDG | 1740 |
| KCLSREEINL LGVVPRLRFI IKNHITLSEA VLELQKNNKI SDYEIEHNEV NLTLPKPSNL | 1800 |
| KFDNGYGGFR NNGEEYVIYN KNTPVPWSNV IANENFGTIV TNNGAGFTYF DSSSEFKITS | 1860 |
| WTNDIVVNDK SEGFKFNGLI FNPEVCVHGI GYSTFGSETI DLKKEITEFV PVNDPVKLYL | 1920 |
| VKLTNKDDSE LEVDVNYWIN PTLGNFEEKT ARHILSEFMG NNNYLKLRNA YSINYGDVCV | 1980 |
| YMSSSEKIDY ALCDKILIKD IGFKLKLGAL QEKTFVFLLS ACRNEELGEA LVRKYSNIDN | 2040 |
| VNKEFRNVKR YWSEKLGVLK VKTCDQSFNY MVNNWYLYQT LSSRIMAKAG FYQVSGAFGY | 2100 |
| RDQLQDAVNI VLVDPDYTRK QILINAEHQF IEGDVLHWWH DRNRFGLRSR YKDDFLWLVY | 2160 |
| ATVIYVNTTG DKSILDEKVP YVSGSILSDY EYEKTMIFDY SSFKETLLEH CLKSLKLSMS | 2220 |
| ALGSHGLPLM GGGDWNDGMN RVGIKGKGES VWLGFFLYSI IDMFVKLVKD NDIYLDISQY | 2280 |
| VSFNKSLKES LNKKAWDGNY YLRAYFDNGD KLGSHENSEC QIDLISQSFS ILSSVIPKNR | 2340 |
| IDKVINNVEE KLVDKDKKII KLLTPAFSKS LDNPGYIMNY PKGIRENGGQ YTHAVSWYIM | 2400 |
| ALIKIGRYDD AYNYFEMINP INRSLNDNDV LKYKVEPYVI AADIYSSSSF PSRGGWTWYT | 2460 |
| GSSGWFYRLA INDILGIKKH SNTLNIVPCI PDDWDGFKAV YKYMNTVYNI EVKRTGIEKI | 2520 |
| TFDGKVVVDI KLVDDNSMHS VIVDVK | 2546 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH94 | 1514 | 2542 | 8.3e-289 | 0.997104247104247 |
| GT84 | 1071 | 1276 | 1.6e-66 | 0.9813953488372092 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3459 | COG3459 | 0.0 | 1514 | 2529 | 2 | 1035 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
| pfam17167 | Glyco_hydro_36 | 1.37e-119 | 2051 | 2478 | 1 | 425 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
| cd11753 | GH94N_ChvB_NdvB_2_like | 2.35e-87 | 1273 | 1597 | 5 | 335 | Second GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This second of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam06165 | Glyco_transf_36 | 1.30e-66 | 1326 | 1571 | 2 | 247 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
| cd11756 | GH94N_ChvB_NdvB_1_like | 2.28e-59 | 1800 | 2036 | 2 | 284 | First GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This first of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCX34529.1 | 0.0 | 10 | 2541 | 70 | 2706 |
| QSZ28298.1 | 0.0 | 10 | 2541 | 72 | 2860 |
| AGB18242.1 | 0.0 | 14 | 2541 | 73 | 2862 |
| ADL68114.1 | 0.0 | 14 | 2541 | 73 | 2862 |
| AST58113.1 | 0.0 | 14 | 2541 | 73 | 2862 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3QDE_A | 1.77e-75 | 1806 | 2524 | 3 | 787 | ChainA, Cellobiose phosphorylase [Acetivibrio thermocellus],3QDE_B Chain B, Cellobiose phosphorylase [Acetivibrio thermocellus] |
| 1V7V_A | 4.32e-74 | 1806 | 2524 | 3 | 778 | Crystalstructure of Vibrio proteolyticus chitobiose phosphorylase [Vibrio proteolyticus],1V7W_A Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc [Vibrio proteolyticus],1V7X_A Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc and sulfate [Vibrio proteolyticus] |
| 3S4C_A | 2.59e-61 | 1806 | 2536 | 3 | 805 | Lactosephosphorylase in complex with sulfate [Cellulomonas uda],3S4D_A Lactose phosphorylase in a ternary complex with cellobiose and sulfate [Cellulomonas uda] |
| 3RRS_A | 1.49e-60 | 1806 | 2536 | 3 | 805 | Crystalstructure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RRS_B Crystal structure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RSY_A Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3RSY_B Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3S4A_A Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4A_B Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4B_A Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda],3S4B_B Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda] |
| 2CQS_A | 3.83e-54 | 1794 | 2524 | 15 | 819 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQS_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQT_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],2CQT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],3QFY_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFY_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFZ_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QFZ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 0.0 | 378 | 2514 | 478 | 2807 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
| Q76IQ9 | 2.12e-73 | 1806 | 2524 | 3 | 778 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio proteolyticus OX=671 GN=chbP PE=1 SV=1 |
| Q9F8X1 | 6.83e-73 | 1806 | 2527 | 3 | 781 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio furnissii OX=29494 GN=chbP PE=1 SV=1 |
| B9K7M6 | 9.88e-71 | 1806 | 2525 | 3 | 790 | Cellobiose phosphorylase OS=Thermotoga neapolitana (strain ATCC 49049 / DSM 4359 / NBRC 107923 / NS-E) OX=309803 GN=cbpA PE=1 SV=1 |
| Q7S0S2 | 3.11e-38 | 2001 | 2521 | 261 | 762 | Cellobionic acid phosphorylase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=NCU09425 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000046 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
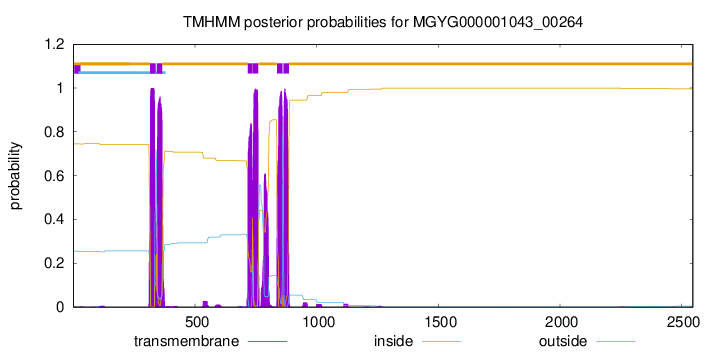
| start | end |
|---|---|
| 316 | 338 |
| 343 | 365 |
| 717 | 736 |
| 738 | 760 |
| 837 | 859 |
| 864 | 886 |
