You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004032_00130
You are here: Home > Sequence: MGYG000004032_00130
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; UMGS1690; | |||||||||||
| CAZyme ID | MGYG000004032_00130 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3742; End: 10254 Strand: - | |||||||||||
Full Sequence Download help
| MDINSKLTFK KPLSLRRKLA KGLSNLRPLY KAAIVNEGTD MSEWLRDNYY LLEREGRSAL | 60 |
| KSLKYSPALP HNGKNIPEIF LLCKKTVGED IEAAIRHEIE NSDRPITTQE LQSLPFMLKA | 120 |
| SLVDTACICA KNRDEYSAEM MGLAITGLRS LPSIDFEQII EDYSQIEKIF SRDPSGAYTG | 180 |
| MDEKTRAIYR SKCTELALRK KCDEAQVAEN IVKLAQSASK SRERHIGFYL LEDTSKSRRI | 240 |
| RGNIFLTLRF IIPLIVSFII SCFSHAYWLI PIIYFPIFET CRPLVDFFAT KGVKPTYLPR | 300 |
| MELKGCIPAS APTLVTISTL IPSPDKAGRL YKRLEQLYRT NCQGEIKFCV LCDLKEYSLP | 360 |
| SKPEDAVSIS AVSRVIKSLN NKYDNKFILV IRPRVEVKTQ RAFAGYERKR GAITELIRYI | 420 |
| RTGVSSFRTI EGDAEFLRRV KYMLALDADT ELLMDSASEM VAAAMHPLNT PVVSESGCVT | 480 |
| NGYGIIAPSV VTSLQSSART PFSVTMAGSG GVTTYDTVSG DIYQDLYGQS IFSGKGLIDT | 540 |
| EAYYRCLDKA FPDERILSHD ILEGSYLRTA YMSDIEMTDE CPSGVLPYLQ RLHRWVRGDW | 600 |
| QNILWLFKRV PSSAGHIVNP INGLSKHMLF DNLRRSATPP FALMCLLLTF FIPSHAALLA | 660 |
| VTALLSAASP GIFAALRSLI GGGVQMLSRK YYSRVMPSAL EALAGALIES IILVQTALIS | 720 |
| LSAICCALYR QFVSHRNLLQ WTTAADAEGS DGFLSSLVFS LPAVIFGVLL FFVNDSFTRL | 780 |
| MACFAIISPL IIYFTSIKRK HETVQIESSE RDTVKAWTAS MWRFYDELAS DQDNFLPPDN | 840 |
| IQESPVHAIA HRTSPTNIGL MMLCTLAARD FGFIDTEHMC VKIEKALCSI EKLEKWNGNL | 900 |
| LNWYDTKTLK PLRPRFVSTV DSGNFACCMS TLSEGLKEYM SESAKISQII DRLENLVDET | 960 |
| DLSPFYNKRR KLFHIGYDLE AEKLSGSYYD LLMSEARMTS YFAIASRQVS KKHWGYLGRT | 1020 |
| LAKQGGYTGP VSWTGTMFEY FMPQLLLPAY ENSLGFEALR FCIHCQRSRA RAKDAPWGCS | 1080 |
| ESAFYAFDPH YNYQYKAHGI QKLGLKRGLN QEYVVSPYSA FLTLPTIPQA SIKNLLRLEE | 1140 |
| MGMTGRYGFY EAADFTKSRA GSGGYSIVRC YMAHHIGMSI LACCNALYNN IMQKRFMNSR | 1200 |
| NMLAAKELLE EKIPAGAAVF DDIAEPEVPQ KPGRTGGEAE IIDDINIAEP KMHLLSNGQY | 1260 |
| TLVVTDSGCG VSFSQGADIY RRSSDILRRP LGLFAVVETK NGAFSITRAP DYYSDAEYSA | 1320 |
| QFAATHAAFY ASKYGLDAGM MICLHPNIPC EERKFIVKNS SSFSQNVSLL LYFEPCLSTP | 1380 |
| EDDSAHPAFS RLFIRCKYIK ELNIITMERR SRKGEPVLYS AAGFIENIPF EYSFNREKVI | 1440 |
| NRPFGAASLI SNMPVLNGDG GVPDPCVAAR VEMNISPKSQ KQFNFVITCA ATPDEAISRL | 1500 |
| VTCRRQGALS SQKAAASPLF DGGLSSRICA SILPKLFYPV KAKGQADKYP VVGGISALWS | 1560 |
| AGISGDFPII LIEVKSADDI SKWEPYINLH SKMRLCGIKF DLCILFSEGG DYMRSVSSAL | 1620 |
| YDIVRLCGSE ELLGQKGGIY CLDVLKLNSE AISAIKAAAS YIAPQNITAS GVSTAKYNPI | 1680 |
| EILPASRPVK PLPSGIEIVG GVFYDSDKGG SFTVTSMPEL PWCQVLANEN FGTLLSDISL | 1740 |
| GYTWAKNSRE NKLTPWFNDT RTDNRGEMLI ARIDGKYYDL IWGSRVTYAR EYALYDGSIG | 1800 |
| GIDYSVKVTV PKHSMTKVIE VNYSRPQGIE TAYYTEPILG VSGTVVKSLS FSFEEEVLNI | 1860 |
| RNGFNTAAPG YMCLSAFGGP VNYVCEKKDF LSGRWDVGQS ASPSNPCGAV IVNQSSTNIV | 1920 |
| FTMSFSETPG YKYDIYLAPE EKAKDITDIK ISTSNKLLDC LINTWLPSQI EQSRIMGRTA | 1980 |
| FYQCGGAFGF RDQLQDVCAQ MIWDPASAKE HILRCCAHQF EEGDVLHWWH PLSSGDAGVR | 2040 |
| TRGSDDMLFL PYTVYEYIDR TADTGILHNI VPFIKGDPLS ENEQERYFVP QISDISGTVY | 2100 |
| EHCLRCVDDK TRCARAPSDR RFRLERRLLQ SWHKGQWRER VACTVCSYCM RAYSIACHAG | 2160 |
| KAKRHRPQTA | 2170 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH94 | 1555 | 2108 | 3.7e-90 | 0.5453667953667953 |
| GT84 | 987 | 1198 | 1.5e-83 | 0.986046511627907 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3459 | COG3459 | 7.40e-53 | 1556 | 2108 | 130 | 722 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
| cd11753 | GH94N_ChvB_NdvB_2_like | 5.11e-42 | 1192 | 1500 | 1 | 321 | Second GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This second of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam06165 | Glyco_transf_36 | 5.32e-29 | 1250 | 1487 | 3 | 246 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
| cd11756 | GH94N_ChvB_NdvB_1_like | 1.63e-27 | 1720 | 1906 | 30 | 227 | First GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This first of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam17167 | Glyco_hydro_36 | 2.62e-26 | 1946 | 2108 | 6 | 171 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI59942.1 | 0.0 | 14 | 2115 | 31 | 2156 |
| CDZ23197.1 | 0.0 | 17 | 2108 | 37 | 2172 |
| AVQ95160.1 | 0.0 | 11 | 2108 | 37 | 2225 |
| AYF37850.1 | 0.0 | 11 | 2108 | 37 | 2225 |
| QCN91406.1 | 0.0 | 11 | 2108 | 37 | 2225 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1V7V_A | 3.59e-09 | 1703 | 2083 | 5 | 464 | Crystalstructure of Vibrio proteolyticus chitobiose phosphorylase [Vibrio proteolyticus],1V7W_A Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc [Vibrio proteolyticus],1V7X_A Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc and sulfate [Vibrio proteolyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 1.07e-216 | 123 | 2108 | 267 | 2509 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
| Q76IQ9 | 1.96e-08 | 1703 | 2083 | 5 | 464 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio proteolyticus OX=671 GN=chbP PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
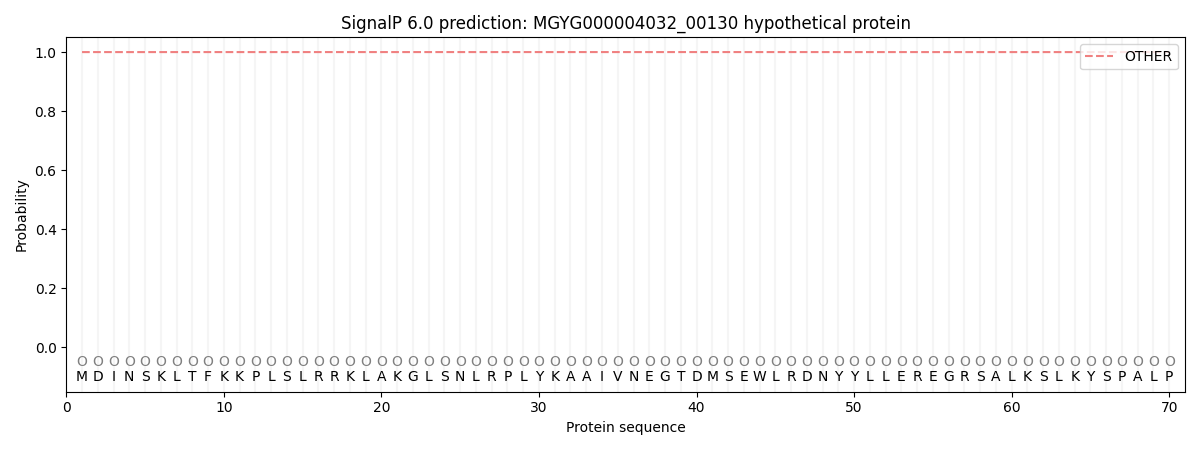
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000080 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
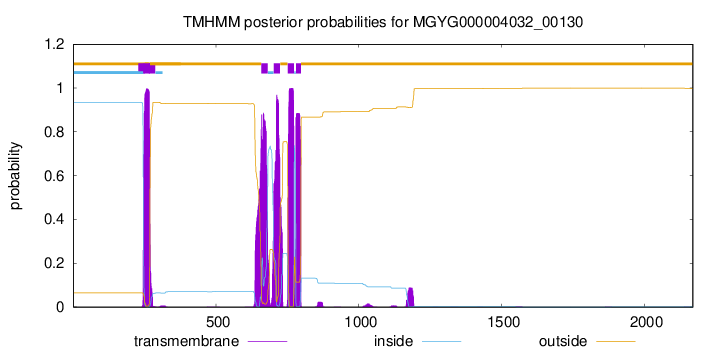
| start | end |
|---|---|
| 246 | 268 |
| 659 | 681 |
| 702 | 724 |
| 751 | 773 |
| 780 | 797 |
