You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000646_00247
You are here: Home > Sequence: MGYG000000646_00247
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Eubacterium_R sp000436835 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; Eubacterium_R; Eubacterium_R sp000436835 | |||||||||||
| CAZyme ID | MGYG000000646_00247 | |||||||||||
| CAZy Family | GH95 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 252961; End: 255627 Strand: - | |||||||||||
Full Sequence Download help
| MNKYIKKILC IVLTVSFAFS LVSCSNGKAD KGVIKSLFTD TSKVSFSYDY TQLKDKKRNK | 60 |
| TKSWRDGMVS GNGLQGFIES GSPYSDTFIF QNMHFIMPNE NQRTCPVTFD ELEKVKQSIV | 120 |
| KGEDITDNSS YDDVYRYHPG GQLRINIAQS RAENYIRYTD YNTSQVGVTY NDKNGIWLRT | 180 |
| SFTSMADGVV VTKLDKSSNG AKLNLTLSYD DLSTLANFGD SDEKNMKYTK LADNSGDYLA | 240 |
| LVSHYPNYEK SELKNGGYAT VTYIITSGGT KERITLDKNI DESQFTGENA GVKITDADCV | 300 |
| YLLTVSERTY DMGDINAFDK QKDFKLVDDC VSTLKNVAQK YNDKSGFNYD LALKNHLAIY | 360 |
| QPQFNNVTLS LGKDNFDSNE KLLKAQKGKK EINSALAERT YYAGRYAYLC CSGYSTSRLY | 420 |
| GMWTGEFNTG WGSKYTMDAN VNLQTSSMNT SNMESTPIGY AYFILRQLPD WEENAYATHG | 480 |
| FTDAIQAPVN TDGDKAVITE TCYPYPFRYW NAGTSWMINP LYETLLSYGN INIPLSDEFN | 540 |
| LDKLKSVLSI SEKDLTDEQI TEIKNRGYLR LEEDILYPLL IKSANYWAQL MTPEYYTAKD | 600 |
| GSIHYEKGKT SLNADETYCI LPSYSPENNP SNYNSPSDAN CAIDISACRD NLEMLKKVMQ | 660 |
| DVDPSADISK WQELEDKLPP YLYDESGALK EWATTAFEEN NEHRHLSHLY GVWPLFETQG | 720 |
| NDKLAKACEQ AIANRTSENE ASHALVHRSL IAARLKDSES LSDALVKLMN HKIRYDSLMT | 780 |
| NHDYDRGSCY CTDFAIGYLG IINEALVYSH DDDIELLPAL PTSGFENGTL KGIKTRNRAT | 840 |
| VTELKWADSG KKISAVITSD IDQTLNVSAK GKTQNVTFKA GESKTIIF | 888 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 53 | 875 | 3.5e-118 | 0.9861495844875346 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO19599.1 | 8.53e-245 | 41 | 888 | 3 | 830 |
| QYR23103.1 | 1.05e-228 | 43 | 846 | 13 | 838 |
| AIQ45759.1 | 2.70e-228 | 63 | 846 | 31 | 839 |
| AIQ51360.1 | 3.55e-227 | 43 | 859 | 13 | 852 |
| AFH60346.1 | 1.20e-225 | 59 | 846 | 27 | 838 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4UFC_A | 1.58e-22 | 158 | 872 | 133 | 750 | Crystalstructure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus],4UFC_B Crystal structure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus] |
| 2RDY_A | 4.97e-17 | 64 | 867 | 14 | 752 | ChainA, BH0842 protein [Halalkalibacterium halodurans C-125],2RDY_B Chain B, BH0842 protein [Halalkalibacterium halodurans C-125] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L7W8 | 5.64e-20 | 61 | 848 | 60 | 794 | Alpha-L-fucosidase 2 OS=Arabidopsis thaliana OX=3702 GN=FUC95A PE=1 SV=1 |
| A2R797 | 6.45e-09 | 300 | 850 | 253 | 760 | Probable alpha-fucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=afcA PE=3 SV=1 |
| Q2USL3 | 5.76e-06 | 652 | 867 | 494 | 717 | Probable alpha-fucosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=afcA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
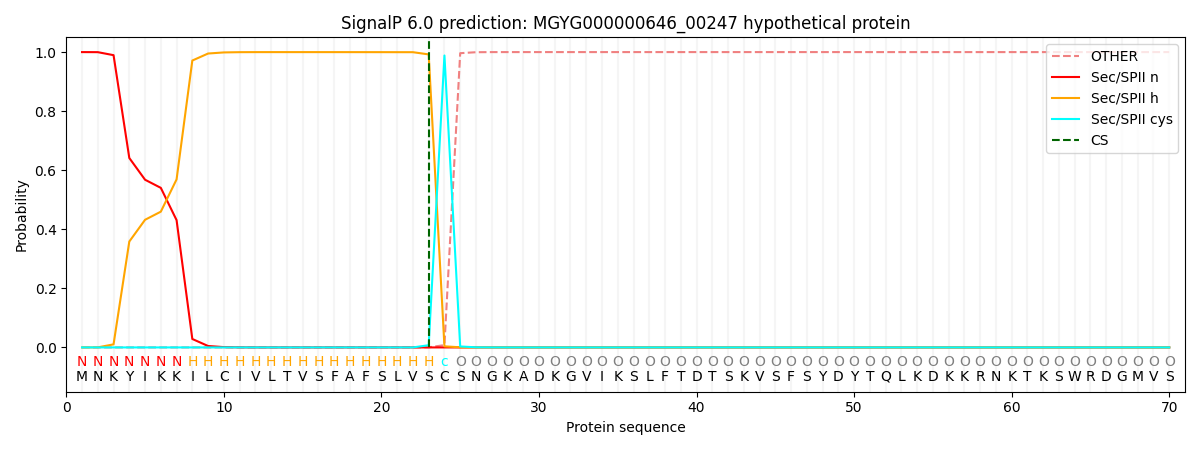
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000038 | 0.999996 | 0.000000 | 0.000000 | 0.000000 |
