You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002262_00024
You are here: Home > Sequence: MGYG000002262_00024
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Eubacterium_R sp900548085 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; Eubacterium_R; Eubacterium_R sp900548085 | |||||||||||
| CAZyme ID | MGYG000002262_00024 | |||||||||||
| CAZy Family | GH95 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29384; End: 32044 Strand: + | |||||||||||
Full Sequence Download help
| MKNLLKKIIS IALCATFTLS FCSCTADKKD LDLKSVFTDT SKVSYTREYV TLKKNKKNKT | 60 |
| NKWRQGMVSG NGLVGYVTSG SPYSDTFIFQ NMHFIMPNEN QRTCPDTSDE LETVKQNIIS | 120 |
| GKDITDDASY DDVYRYHPGG QLRVESEKHH EKDYVSYTDY ETSQTGVRYT DTDGTWERKS | 180 |
| FTSMADGVSI TELTASSSGK KATVTLSYDD ISTLANFGDS DEVNLRYKKV TASDGSYLGI | 240 |
| VAHYPDYKNS ELKNGGYATV TYIVTDGKKE KISLDKKTDE SQFFGENTGV KVSGADSIYL | 300 |
| ITVSDRTYDM GAYSDFDKTE EYPLVSDCVS TLSAVAKKYG TSGSFDYKTA LAEHLKLYQP | 360 |
| QFDAVTLTLD DDNTSSNEAL LSEQKGKKEI NSALAQRTYY AGRYAYLCCS GYSTSRLYGM | 420 |
| WTGEWNTGWG SKYTMDANVN LQTSSMNTGN MSSTPIGYTY FILRQLPDWE ENARATHGYT | 480 |
| DAIQAPVNTD GDKAVITETC YPYPFRYWNA GTSWMIQPLY ETLQTYGNIN IPLSDEFDLN | 540 |
| ELKSVLSPTE KDLSDEDILA VKKRGYLRLE EDVLYPLLIK SANYWAQLLT PEYYTAADGS | 600 |
| IHYEKGKKEL NSDEKYCILP SYSPENNPKN YPSPSDANCA IDISACRDNI DMLLAVSEEV | 660 |
| SPETDVTRWQ ELKNNLPPYL YDDTGALKEW ATTAFQENNE HRHLSHLYGV WPLFETQNNP | 720 |
| GLTKACEQAI ANRKSENEAS HALVHRSLIS ARLKDSASLT DALLNLMNHK IRYNSLMTNH | 780 |
| DYDRGSCYCT DFVIGYLGIV NEALVYSHDS DIGLLPALPD SGFDSGTLTG VRTRNRAVVK | 840 |
| KLQWDNHTKK VTAEITSDID QTLNISYGKA KKTLAFKAGE TKTIEF | 886 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 57 | 871 | 3.5e-119 | 0.9764542936288089 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO19599.1 | 3.62e-243 | 63 | 886 | 27 | 830 |
| ACB76390.1 | 1.36e-234 | 63 | 879 | 62 | 902 |
| AIQ45759.1 | 5.56e-233 | 61 | 845 | 30 | 840 |
| AIQ51360.1 | 1.03e-231 | 58 | 857 | 27 | 852 |
| QYR23103.1 | 1.39e-228 | 63 | 860 | 32 | 854 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4UFC_A | 8.93e-20 | 347 | 865 | 281 | 741 | Crystalstructure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus],4UFC_B Crystal structure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus] |
| 2RDY_A | 4.17e-18 | 286 | 865 | 223 | 752 | ChainA, BH0842 protein [Halalkalibacterium halodurans C-125],2RDY_B Chain B, BH0842 protein [Halalkalibacterium halodurans C-125] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L7W8 | 9.96e-15 | 310 | 844 | 301 | 792 | Alpha-L-fucosidase 2 OS=Arabidopsis thaliana OX=3702 GN=FUC95A PE=1 SV=1 |
| A2R797 | 3.76e-14 | 290 | 846 | 244 | 758 | Probable alpha-fucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=afcA PE=3 SV=1 |
| Q2USL3 | 1.11e-06 | 659 | 846 | 484 | 696 | Probable alpha-fucosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=afcA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
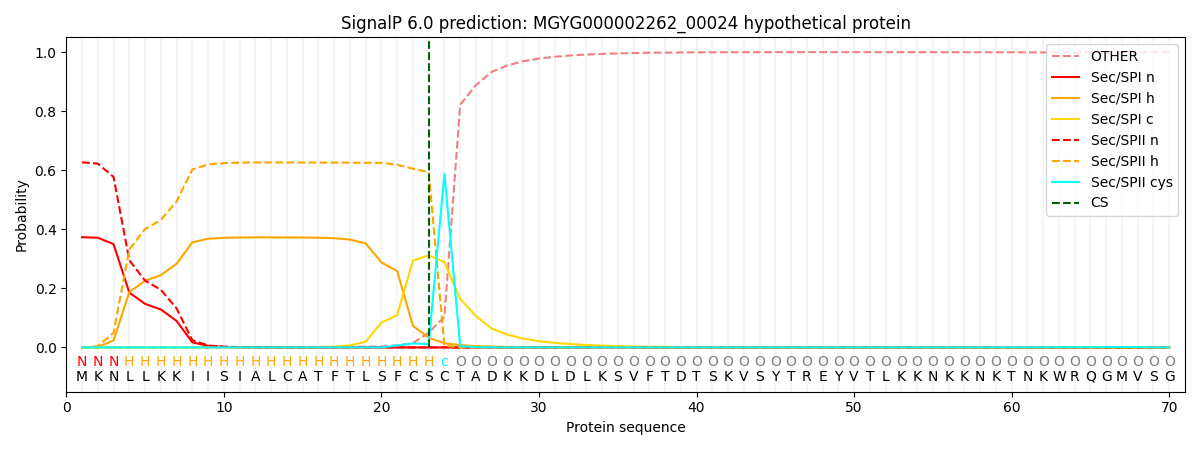
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000403 | 0.366019 | 0.633122 | 0.000138 | 0.000168 | 0.000146 |
