You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002603_00183
You are here: Home > Sequence: MGYG000002603_00183
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900767615 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900767615 | |||||||||||
| CAZyme ID | MGYG000002603_00183 | |||||||||||
| CAZy Family | GH95 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 224563; End: 227019 Strand: + | |||||||||||
Full Sequence Download help
| MSKKITTILT FAMMGALQAS AQQQTQLSYW FDTPVTLKGQ QIWYGGHPEK WTNKKPISAG | 60 |
| DTARNPDPDW ESKSLPIGNG SIGANILGSV EAERITLNEK TLWRGGPNTK KGAAYYWNVN | 120 |
| KNSAHVMQEI RDAFAANDWE KASQLTRKNF NSPVPYESYA EDPFRFGSFT TMGEAYIETG | 180 |
| LSTVGMSKYR RALSLDSALA SVSFVKDGVR YQREYFISYP ANVMAIRYKA SRQGKQNLVF | 240 |
| SYAPNPVSTG RMVADGRNGL LWDARLDNND MQYAIRIRAI NKGGSLSNEG GKLTVKGADE | 300 |
| VVFLISADTD YKANNNPDYN DSKTYVGVDP LATTQDWVSK AEGKGYVQLL DEHYKDYSRL | 360 |
| FNRVSLNLNN VDKANDMPID QRLANYRKGA KDFYLEQLYY QFGRYLLIAS SRPGNMPANL | 420 |
| QGVWHNNVDG PWRVDYHNNI NLQMNYWPAC TTNLSECELP LFDFIQTQVK PGEKTAKSYY | 480 |
| GTRGWTTSVS SNIFGFTSPL ASEDMSWNFS PFAGPWLATH LWDYYDYTRD KKFLSETAYD | 540 |
| IIKGSANFAA DYLWHRKDGV YTAAPSTSPE HGPIDEGATF AHAVIREILL DAVEASKILG | 600 |
| KDAKDRKLWE DALKHIAPYQ IGRYGQLMEW SKDIDDPKDE HRHVNHLFGL HPGRTISPIT | 660 |
| TPELAKASKV VLEHRGDGAT GWSMGWKLNQ WARLHDGNHA YKLYGNLLKN GTLDNLWDTH | 720 |
| APFQIDGNFG GTAGVTEMLM QSHMGFIHLL PALPDAWQKG SIKGLRAKGN FTVDIYWDNG | 780 |
| RLTKAVILSG SGEPCQVRYG DKVKKMKTQK GKTYEVKF | 818 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 63 | 806 | 5.6e-273 | 0.981994459833795 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14498 | Glyco_hyd_65N_2 | 1.21e-57 | 65 | 313 | 5 | 233 | Glycosyl hydrolase family 65, N-terminal domain. This domain represents a domain found to the N-terminus of the glycosyl hydrolase 65 family catalytic domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKH85901.1 | 0.0 | 3 | 816 | 2 | 819 |
| QCQ35127.1 | 0.0 | 3 | 816 | 2 | 819 |
| QCQ48529.1 | 0.0 | 3 | 816 | 2 | 819 |
| QTO26524.1 | 0.0 | 3 | 816 | 2 | 819 |
| QLK81254.1 | 0.0 | 3 | 816 | 2 | 819 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2RDY_A | 1.05e-165 | 72 | 797 | 15 | 752 | ChainA, BH0842 protein [Halalkalibacterium halodurans C-125],2RDY_B Chain B, BH0842 protein [Halalkalibacterium halodurans C-125] |
| 4UFC_A | 9.40e-159 | 66 | 798 | 29 | 742 | Crystalstructure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus],4UFC_B Crystal structure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus] |
| 7KMQ_A | 2.70e-150 | 50 | 799 | 28 | 759 | ChainA, Glyco_hyd_65N_2 domain-containing protein [Xanthomonas citri pv. citri str. 306],7KMQ_B Chain B, Glyco_hyd_65N_2 domain-containing protein [Xanthomonas citri pv. citri str. 306] |
| 2EAB_A | 3.74e-137 | 36 | 798 | 14 | 850 | Crystalstructure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAB_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAC_A Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum],2EAC_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum] |
| 2EAD_A | 2.85e-136 | 36 | 798 | 14 | 850 | ChainA, Alpha-fucosidase [Bifidobacterium bifidum],2EAD_B Chain B, Alpha-fucosidase [Bifidobacterium bifidum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L7W8 | 3.46e-137 | 66 | 784 | 59 | 799 | Alpha-L-fucosidase 2 OS=Arabidopsis thaliana OX=3702 GN=FUC95A PE=1 SV=1 |
| A2R797 | 1.72e-103 | 74 | 799 | 38 | 781 | Probable alpha-fucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=afcA PE=3 SV=1 |
| Q5AU81 | 7.28e-83 | 77 | 778 | 46 | 776 | Alpha-fucosidase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=afcA PE=1 SV=1 |
| Q2USL3 | 4.52e-70 | 69 | 798 | 27 | 713 | Probable alpha-fucosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=afcA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
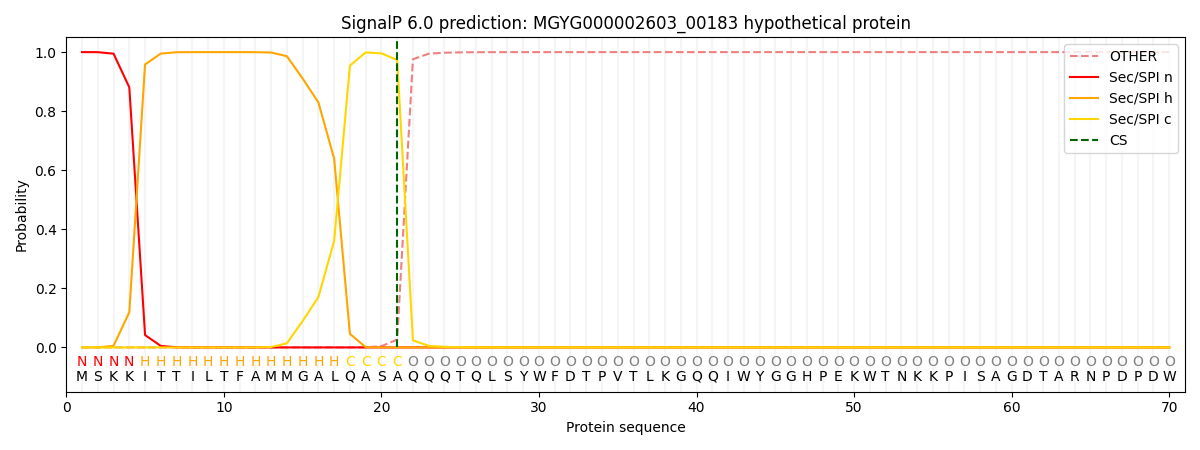
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000267 | 0.999085 | 0.000174 | 0.000163 | 0.000145 | 0.000134 |
