You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000091_00400
You are here: Home > Sequence: MGYG000000091_00400
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
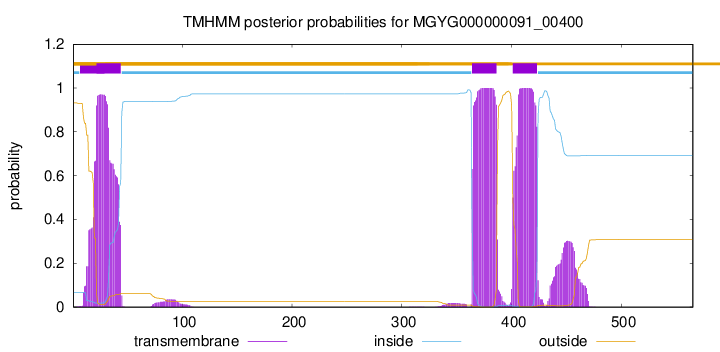
TMHMM annotations
Basic Information help
| Species | Achromobacter xylosoxidans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Burkholderiaceae; Achromobacter; Achromobacter xylosoxidans | |||||||||||
| CAZyme ID | MGYG000000091_00400 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 468112; End: 469809 Strand: - | |||||||||||
Full Sequence Download help
| MSSLYWPYLI ADYYRGLEIV TAVVGVIILL SSLDDLFIDG WYWVRELRRA LTVKRRYTPL | 60 |
| TAAQLRAKEE QPLAIMVPAW LEYDVIASML ETMVSTLEYK NYMIFAGTYQ NDERTIKEVE | 120 |
| RMRRRYRQLI RVEVPHDGPT CKADCLNWIV QAIFAQDARQ PVPFAGVILH DSEDVLHPLE | 180 |
| LKYYNYLLPR IDFIQLPVTS LERHWYELVA GTYMDEFAEW HTKDLVVRES LSKMVPSAGV | 240 |
| GTCFSRRALV ELAAETDNQP FNTDTLTEDY DIGARLAQRG MRQIFGKFGV EYVTRRRAWF | 300 |
| GLGKEKVSSI HMPLGVREYF PNTFRTAYRQ KARWTLGIGL QGWQQVGWTG SLATKYLLFR | 360 |
| DRKGLVTSFI AILAYLLLAN FFLFYLADLF GWWTVYYPSY FRPGGWLVTL MWINAVALLL | 420 |
| RVVQRAYFVG RMYGWEHAIL SIPRMIVGNF INAMAAARAW KLFIGHLITG KRLAWDKTMH | 480 |
| DFPSTDQLAQ QRQRLGELLL SWQAIDEDKL ARALEIQSRD KRPLGEILME QGWLDEGTLR | 540 |
| EAIHFQQGGQ ADTAPTHGEN KPATP | 565 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 167 | 386 | 1.7e-49 | 0.9796954314720813 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK11234 | nfrB | 0.0 | 9 | 548 | 1 | 536 | phage adsorption protein NrfB. |
| PRK14716 | PRK14716 | 0.0 | 3 | 519 | 1 | 504 | glycosyl transferase family protein. |
| PRK15489 | nfrB | 0.0 | 1 | 546 | 1 | 544 | glycosyl transferase family protein. |
| pfam13632 | Glyco_trans_2_3 | 2.48e-35 | 167 | 386 | 2 | 193 | Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis. |
| pfam13641 | Glyco_tranf_2_3 | 5.79e-25 | 73 | 337 | 4 | 230 | Glycosyltransferase like family 2. Members of this family of prokaryotic proteins include putative glucosyltransferase, which are involved in bacterial capsule biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQE57003.1 | 0.0 | 1 | 565 | 1 | 565 |
| QKI71954.1 | 0.0 | 1 | 565 | 1 | 565 |
| QKQ56310.1 | 0.0 | 1 | 565 | 1 | 565 |
| QNP84402.1 | 0.0 | 1 | 565 | 1 | 559 |
| QEK94389.1 | 0.0 | 1 | 555 | 1 | 555 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0AFA5 | 1.68e-133 | 11 | 514 | 9 | 509 | Bacteriophage adsorption protein B OS=Escherichia coli (strain K12) OX=83333 GN=nfrB PE=4 SV=1 |
| P0AFA6 | 1.68e-133 | 11 | 514 | 9 | 509 | Bacteriophage N4 adsorption protein B OS=Escherichia coli O157:H7 OX=83334 GN=nfrB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
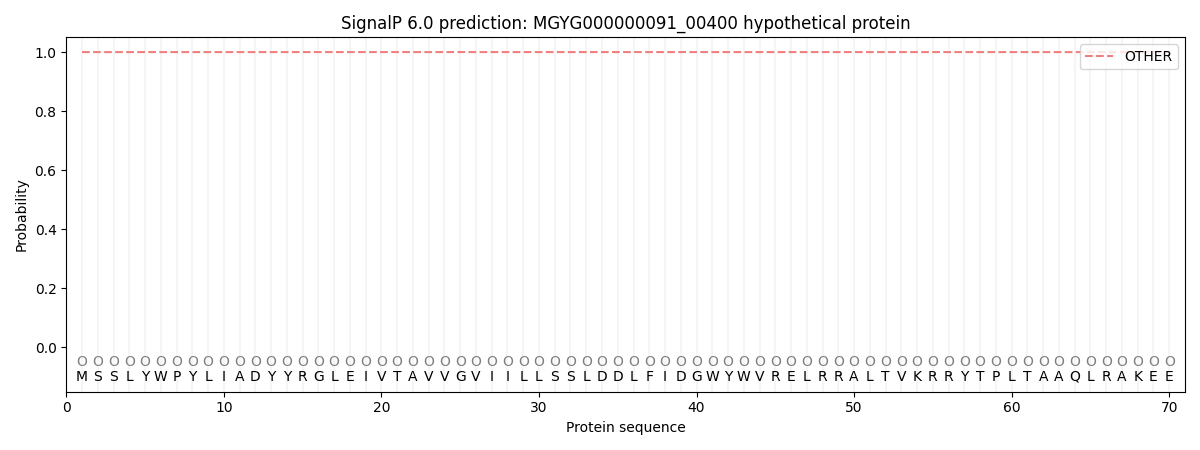
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000054 | 0.000006 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |