You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002481_00936
You are here: Home > Sequence: MGYG000002481_00936
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Stenotrophomonas maltophilia_F | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Stenotrophomonas; Stenotrophomonas maltophilia_F | |||||||||||
| CAZyme ID | MGYG000002481_00936 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8739; End: 9878 Strand: - | |||||||||||
Full Sequence Download help
| MKTLLFLAMC AFILVRVCVQ ASRRPSTRLY SIDAIVPAYN EAPCLERSLT GLLQNPYVAR | 60 |
| VICVNDGSTD NTAQVLDALQ ARWPGRLVAV HQANTGKGGA LMHGLRHATA EQVFLTDADT | 120 |
| HIDPRGHGLG HLLDEIERGA DAVGGVPSSS LQGAGFLPHV RASLKLPMIV IKRSFQQWLG | 180 |
| GAPFIVSGSC GLFRTSVLRK VGFSDRTRVE DLDMSWSLVA QGYRVRQSVR CVVYPQECNT | 240 |
| LVEEWRRWRR WIVGYAVCMR LHRGLLLTRF GVFSILPMML LALAGIVLTL QVWFGAAQLH | 300 |
| GPGGLALSVM PLFWIGIVML LAVISAIHHR RASLVPAAAF SMLYVLLAYA IWLMHGVAGL | 360 |
| FTGREPARDK PTRYPHVVE | 379 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 34 | 201 | 1.1e-28 | 0.9823529411764705 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1215 | BcsA | 2.26e-42 | 4 | 352 | 28 | 392 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| cd06423 | CESA_like | 1.34e-31 | 35 | 211 | 2 | 180 | CESA_like is the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of Glucan. |
| PRK11204 | PRK11204 | 1.04e-30 | 35 | 322 | 59 | 352 | N-glycosyltransferase; Provisional |
| pfam00535 | Glycos_transf_2 | 3.97e-29 | 33 | 198 | 1 | 164 | Glycosyl transferase family 2. Diverse family, transferring sugar from UDP-glucose, UDP-N-acetyl- galactosamine, GDP-mannose or CDP-abequose, to a range of substrates including cellulose, dolichol phosphate and teichoic acids. |
| cd04179 | DPM_DPG-synthase_like | 4.57e-27 | 35 | 201 | 2 | 166 | DPM_DPG-synthase_like is a member of the Glycosyltransferase 2 superfamily. DPM1 is the catalytic subunit of eukaryotic dolichol-phosphate mannose (DPM) synthase. DPM synthase is required for synthesis of the glycosylphosphatidylinositol (GPI) anchor, N-glycan precursor, protein O-mannose, and C-mannose. In higher eukaryotes,the enzyme has three subunits, DPM1, DPM2 and DPM3. DPM is synthesized from dolichol phosphate and GDP-Man on the cytosolic surface of the ER membrane by DPM synthase and then is flipped onto the luminal side and used as a donor substrate. In lower eukaryotes, such as Saccharomyces cerevisiae and Trypanosoma brucei, DPM synthase consists of a single component (Dpm1p and TbDpm1, respectively) that possesses one predicted transmembrane region near the C terminus for anchoring to the ER membrane. In contrast, the Dpm1 homologues of higher eukaryotes, namely fission yeast, fungi, and animals, have no transmembrane region, suggesting the existence of adapter molecules for membrane anchoring. This family also includes bacteria and archaea DPM1_like enzymes. However, the enzyme structure and mechanism of function are not well understood. The UDP-glucose:dolichyl-phosphate glucosyltransferase (DPG_synthase) is a transmembrane-bound enzyme of the endoplasmic reticulum involved in protein N-linked glycosylation. This enzyme catalyzes the transfer of glucose from UDP-glucose to dolichyl phosphate. This protein family belongs to Glycosyltransferase 2 superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCB33872.1 | 1.23e-277 | 1 | 379 | 1 | 379 |
| QGL82844.1 | 1.23e-277 | 1 | 379 | 1 | 379 |
| QNG84847.1 | 1.23e-277 | 1 | 379 | 1 | 379 |
| QNG89483.1 | 1.23e-277 | 1 | 379 | 1 | 379 |
| AWB76649.1 | 1.23e-277 | 1 | 379 | 1 | 379 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6YV7_B | 5.35e-08 | 32 | 224 | 44 | 235 | MannosyltransferasePcManGT from Pyrobaculum calidifontis [Pyrobaculum calidifontis JCM 11548],6YV8_B Mannosyltransferase PcManGT from Pyrobaculum calidifontis in complex with GDP and Mn2+ [Pyrobaculum calidifontis JCM 11548],6YV9_A Mannosyltransferase PcManGT from Pyrobaculum calidifontis in complex with GDP-Man and Mn2+ [Pyrobaculum calidifontis JCM 11548] |
| 6YV7_A | 5.37e-08 | 32 | 224 | 45 | 236 | MannosyltransferasePcManGT from Pyrobaculum calidifontis [Pyrobaculum calidifontis JCM 11548],6YV8_A Mannosyltransferase PcManGT from Pyrobaculum calidifontis in complex with GDP and Mn2+ [Pyrobaculum calidifontis JCM 11548],6YV9_B Mannosyltransferase PcManGT from Pyrobaculum calidifontis in complex with GDP-Man and Mn2+ [Pyrobaculum calidifontis JCM 11548] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q47536 | 1.75e-169 | 1 | 379 | 1 | 379 | Uncharacterized glycosyltransferase YaiP OS=Escherichia coli (strain K12) OX=83333 GN=yaiP PE=3 SV=2 |
| Q8GLC5 | 6.42e-17 | 32 | 254 | 49 | 270 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus epidermidis OX=1282 GN=icaA PE=3 SV=1 |
| Q5HKQ0 | 8.65e-17 | 32 | 254 | 49 | 270 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=icaA PE=1 SV=1 |
| Q5HCN1 | 3.83e-16 | 29 | 254 | 46 | 270 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus aureus (strain COL) OX=93062 GN=icaA PE=3 SV=1 |
| Q6GDD8 | 3.83e-16 | 29 | 254 | 46 | 270 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=icaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
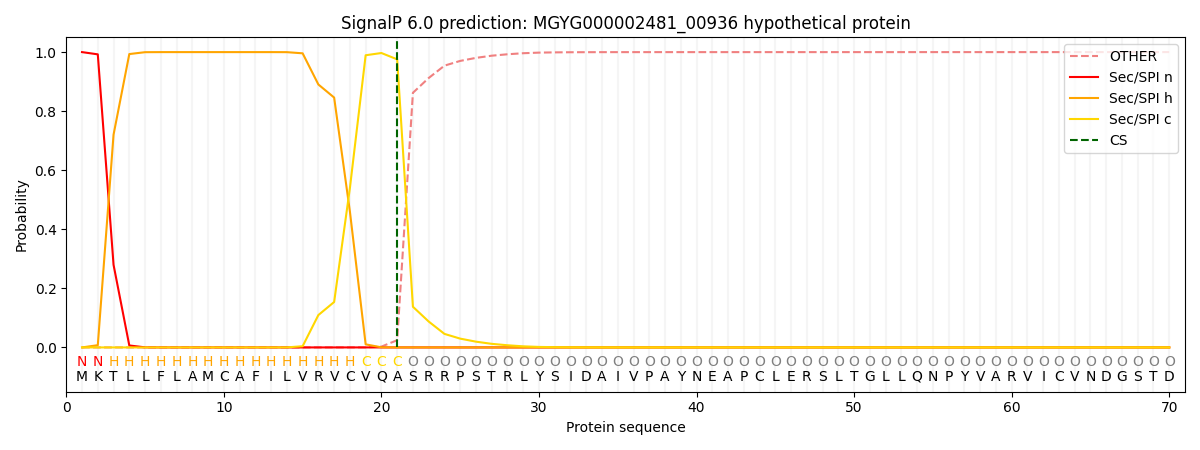
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000248 | 0.999152 | 0.000149 | 0.000133 | 0.000124 | 0.000126 |
TMHMM Annotations download full data without filtering help
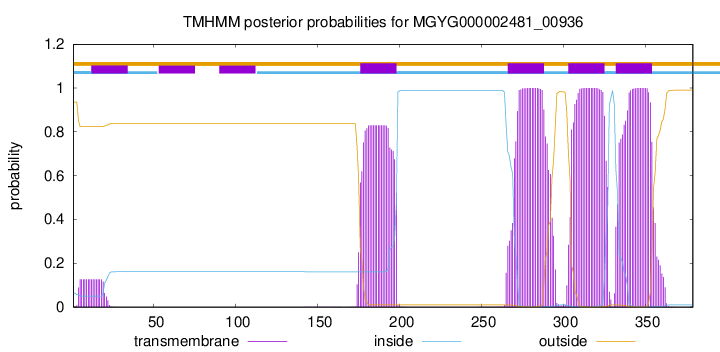
| start | end |
|---|---|
| 176 | 198 |
| 266 | 288 |
| 303 | 325 |
| 332 | 354 |
