You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003630_00594
You are here: Home > Sequence: MGYG000003630_00594
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | F082 sp900771045 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; F082; F082; F082 sp900771045 | |||||||||||
| CAZyme ID | MGYG000003630_00594 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8882; End: 10822 Strand: - | |||||||||||
Full Sequence Download help
| MKLSVVIVNY NVEHFLEQCL LSVRRAMRGV DGEVFVVDNN SVDGSLKMLA AKFPEVKVIA | 60 |
| NKENVGFSRA NNQAIRVSGG EYVLLLNPDT VVEDDTFVKV LDFMDSHPDA GGLGVKMVDG | 120 |
| KGCFLPESKR GLPTPMTAFY KIFGLSKLFP KSKRFGRYHL GFLDENETNE VDVLSGAFML | 180 |
| LRRAALDKSG LLDETFFMYG EDIDMSYRIT LAGYKNYYFH DTRIIHYKGE STKKTSVNYV | 240 |
| IVFYKAMEIF AKKHFAKDGA WFLSFLINTA IYLRAGAAIF SRFLSCVAQP LTEAVTGFAG | 300 |
| LAVISGLWGH FSVYDGIGNY PRMLFTIVIP LYILIWIASS YVIGGYDRPY RKTPSMVGNV | 360 |
| VGMLVILVIY ALLPEEMRFS RALILFGTAW MCVVSLLTRG IGRLIGLANY KPEHSKHRFL | 420 |
| VIGNADEVRR VATLLESTAI KPDYICNVLP DEDAELPEGY VGCLRQVPDI AAIYKIDEVI | 480 |
| FCAKDVPNAV IIDKMVAWQE LGLDYKIAPE DALSIIGSNS INTRGDLYTV TVNTIVTVRN | 540 |
| KRRKRLFDLS VACIVIFSWP LTAWFVKNPV SLLKNGVRVL FGRRTWVGFC SSDTRLPILK | 600 |
| KHIFTTAMLL KNKKIDSKTA YEIDVRYAQD YKLVTDLNIF LEAVKK | 646 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 4 | 188 | 4.6e-24 | 0.9823529411764705 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd04186 | GT_2_like_c | 9.85e-56 | 5 | 227 | 1 | 165 | Subfamily of Glycosyltransferase Family GT2 of unknown function. GT-2 includes diverse families of glycosyltransferases with a common GT-A type structural fold, which has two tightly associated beta/alpha/beta domains that tend to form a continuous central sheet of at least eight beta-strands. These are enzymes that catalyze the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. Glycosyltransferases have been classified into more than 90 distinct sequence based families. |
| COG1216 | GT2 | 6.82e-52 | 1 | 284 | 3 | 279 | Glycosyltransferase, GT2 family [Carbohydrate transport and metabolism]. |
| pfam00535 | Glycos_transf_2 | 4.60e-24 | 4 | 186 | 1 | 164 | Glycosyl transferase family 2. Diverse family, transferring sugar from UDP-glucose, UDP-N-acetyl- galactosamine, GDP-mannose or CDP-abequose, to a range of substrates including cellulose, dolichol phosphate and teichoic acids. |
| cd02525 | Succinoglycan_BP_ExoA | 1.56e-20 | 4 | 239 | 3 | 220 | ExoA is involved in the biosynthesis of succinoglycan. Succinoglycan Biosynthesis Protein ExoA catalyzes the formation of a beta-1,3 linkage of the second sugar (glucose) of the succinoglycan with the galactose on the lipid carrie. Succinoglycan is an acidic exopolysaccharide that is important for invasion of the nodules. Succinoglycan is a high-molecular-weight polymer composed of repeating octasaccharide units. These units are synthesized on membrane-bound isoprenoid lipid carriers, beginning with galactose followed by seven glucose molecules, and modified by the addition of acetate, succinate, and pyruvate. ExoA is a membrane protein with a transmembrance domain at c-terminus. |
| pfam13641 | Glyco_tranf_2_3 | 2.92e-18 | 3 | 235 | 4 | 220 | Glycosyltransferase like family 2. Members of this family of prokaryotic proteins include putative glucosyltransferase, which are involved in bacterial capsule biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGY45414.1 | 3.75e-218 | 1 | 645 | 1 | 646 |
| QIA08556.1 | 1.25e-213 | 1 | 646 | 1 | 648 |
| AHW61351.1 | 2.87e-212 | 1 | 645 | 1 | 647 |
| AEV32005.1 | 2.75e-210 | 1 | 645 | 1 | 648 |
| ASS48316.1 | 8.92e-209 | 2 | 646 | 7 | 647 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6P61_A | 3.14e-06 | 4 | 125 | 16 | 140 | Structureof a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_B Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_C Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_D Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197] |
| 5MLZ_A | 9.40e-06 | 1 | 120 | 23 | 143 | Dolichylphosphate mannose synthase in complex with GDP and Mg2+ [Pyrococcus furiosus DSM 3638],5MM0_A Dolichyl phosphate mannose synthase in complex with GDP-mannose and Mn2+ (donor complex) [Pyrococcus furiosus DSM 3638],5MM1_A Dolichyl phosphate mannose synthase in complex with GDP and dolichyl phosphate mannose [Pyrococcus furiosus DSM 3638] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WMY2 | 8.90e-20 | 3 | 261 | 5 | 268 | N-acetylglucosaminyl-diphospho-decaprenol L-rhamnosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=wbbL PE=3 SV=2 |
| P9WMY3 | 8.90e-20 | 3 | 261 | 5 | 268 | N-acetylglucosaminyl-diphospho-decaprenol L-rhamnosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=wbbL PE=1 SV=2 |
| P46918 | 1.85e-09 | 4 | 107 | 11 | 118 | Minor teichoic acid biosynthesis protein GgaB OS=Bacillus subtilis (strain 168) OX=224308 GN=ggaB PE=3 SV=1 |
| D4GU63 | 4.06e-09 | 3 | 208 | 20 | 205 | Low-salt glycan biosynthesis hexosyltransferase Agl10 OS=Haloferax volcanii (strain ATCC 29605 / DSM 3757 / JCM 8879 / NBRC 14742 / NCIMB 2012 / VKM B-1768 / DS2) OX=309800 GN=agl10 PE=3 SV=1 |
| E0U4V7 | 3.22e-08 | 1 | 128 | 1 | 132 | Poly(ribitol-phosphate) beta-glucosyltransferase OS=Bacillus spizizenii (strain ATCC 23059 / NRRL B-14472 / W23) OX=655816 GN=tarQ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000046 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
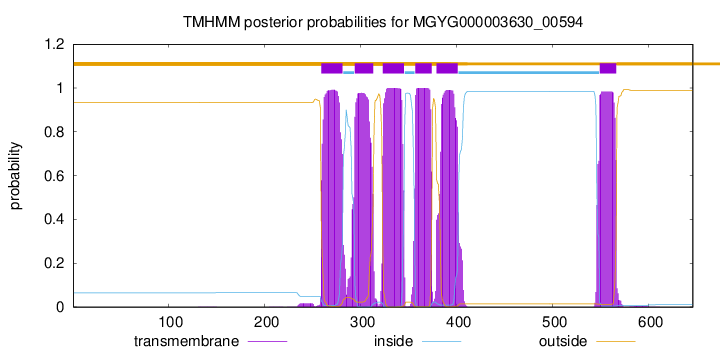
| start | end |
|---|---|
| 259 | 281 |
| 294 | 313 |
| 323 | 345 |
| 357 | 374 |
| 379 | 401 |
| 549 | 566 |
