You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004285_00444
You are here: Home > Sequence: MGYG000004285_00444
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS1670 sp900546215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Anaerotignaceae; UMGS1670; UMGS1670 sp900546215 | |||||||||||
| CAZyme ID | MGYG000004285_00444 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20515; End: 21924 Strand: + | |||||||||||
Full Sequence Download help
| MTKAVSLFLD GVGIFFLTYL VIYASYLFLS VAMGAWNLYK RDKMLLIKNE LKHDFYFPVS | 60 |
| ILVPAYNEEV TILDSIKSLF NLDYRLYEII VIDDGSTDKT AETVISFFDM HKVSKPIRLV | 120 |
| LKCQEIEAVY ETELDNIRLT LICKKNGGKG DALNVGINAS QYPYFLCIDA DSMLQRDSLE | 180 |
| RIVQPVMEDD GVIAVGGLIR IAQCISLENG NAVGYHLPWN PLLSMQVMEY DRSFLASRIL | 240 |
| LDAFNGNLII SGAFGLFKKD IAVAVGGYDT YTLGEDMELV MKLHTFSRNN EMKYSIRYEP | 300 |
| NAVCWSQAPS SMGDLAKQRR RWHLGLFQSM MKYRSMFLNP RFGLVGFFSY IYYLIYELFT | 360 |
| PVIEIFGIIT IIIAGIFGLL NIPFMIQFYV LYVVYGAVLT ITAFFQRIYT QNLKISGSDI | 420 |
| LRAVVICFLE SMFFRFVLAF IRSTALLRYS QYKNNWDKIK RERHSEIKL | 469 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 60 | 202 | 1.1e-22 | 0.7 |
| GT2 | 164 | 374 | 1.7e-17 | 0.949238578680203 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06423 | CESA_like | 8.92e-60 | 61 | 276 | 1 | 180 | CESA_like is the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of Glucan. |
| COG1215 | BcsA | 1.19e-50 | 9 | 462 | 9 | 425 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| PRK11204 | PRK11204 | 8.27e-45 | 9 | 341 | 3 | 294 | N-glycosyltransferase; Provisional |
| cd02525 | Succinoglycan_BP_ExoA | 6.71e-27 | 59 | 348 | 2 | 248 | ExoA is involved in the biosynthesis of succinoglycan. Succinoglycan Biosynthesis Protein ExoA catalyzes the formation of a beta-1,3 linkage of the second sugar (glucose) of the succinoglycan with the galactose on the lipid carrie. Succinoglycan is an acidic exopolysaccharide that is important for invasion of the nodules. Succinoglycan is a high-molecular-weight polymer composed of repeating octasaccharide units. These units are synthesized on membrane-bound isoprenoid lipid carriers, beginning with galactose followed by seven glucose molecules, and modified by the addition of acetate, succinate, and pyruvate. ExoA is a membrane protein with a transmembrance domain at c-terminus. |
| PRK14583 | hmsR | 6.28e-26 | 59 | 355 | 77 | 341 | poly-beta-1,6 N-acetyl-D-glucosamine synthase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMT19152.1 | 5.53e-184 | 1 | 464 | 1 | 464 |
| QNK56534.1 | 2.09e-183 | 1 | 463 | 1 | 463 |
| QBE99872.1 | 3.45e-175 | 1 | 463 | 1 | 463 |
| QIB55150.1 | 4.89e-175 | 1 | 463 | 1 | 463 |
| QMW76995.1 | 4.89e-175 | 1 | 463 | 1 | 463 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5HEA_A | 1.84e-08 | 59 | 195 | 7 | 118 | CgTstructure in hexamer [Streptococcus parasanguinis FW213],5HEA_B CgT structure in hexamer [Streptococcus parasanguinis FW213],5HEA_C CgT structure in hexamer [Streptococcus parasanguinis FW213],5HEC_A CgT structure in dimer [Streptococcus parasanguinis FW213],5HEC_B CgT structure in dimer [Streptococcus parasanguinis FW213] |
| 5TZE_C | 3.29e-06 | 60 | 180 | 4 | 102 | Crystalstructure of S. aureus TarS in complex with UDP-GlcNAc [Staphylococcus aureus],5TZE_E Crystal structure of S. aureus TarS in complex with UDP-GlcNAc [Staphylococcus aureus],5TZI_C Crystal structure of S. aureus TarS 1-349 [Staphylococcus aureus],5TZJ_A Crystal structure of S. aureus TarS 1-349 in complex with UDP-GlcNAc [Staphylococcus aureus],5TZJ_C Crystal structure of S. aureus TarS 1-349 in complex with UDP-GlcNAc [Staphylococcus aureus],5TZK_C Crystal structure of S. aureus TarS 1-349 in complex with UDP [Staphylococcus aureus] |
| 5TZ8_A | 4.46e-06 | 60 | 180 | 4 | 102 | Crystalstructure of S. aureus TarS [Staphylococcus aureus],5TZ8_B Crystal structure of S. aureus TarS [Staphylococcus aureus],5TZ8_C Crystal structure of S. aureus TarS [Staphylococcus aureus] |
| 6P61_A | 9.00e-06 | 59 | 177 | 15 | 108 | Structureof a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_B Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_C Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_D Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8GLC5 | 8.25e-33 | 42 | 332 | 34 | 277 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus epidermidis OX=1282 GN=icaA PE=3 SV=1 |
| Q5HKQ0 | 2.14e-32 | 42 | 332 | 34 | 277 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=icaA PE=1 SV=1 |
| Q7A351 | 3.39e-30 | 48 | 332 | 38 | 277 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus aureus (strain N315) OX=158879 GN=icaA PE=3 SV=1 |
| Q5HCN1 | 3.39e-30 | 48 | 332 | 38 | 277 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus aureus (strain COL) OX=93062 GN=icaA PE=3 SV=1 |
| Q6GDD8 | 3.39e-30 | 48 | 332 | 38 | 277 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=icaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999901 | 0.000127 | 0.000001 | 0.000001 | 0.000000 | 0.000008 |
TMHMM Annotations download full data without filtering help
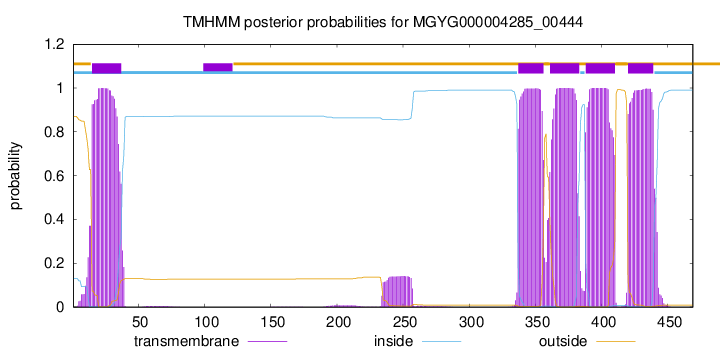
| start | end |
|---|---|
| 15 | 37 |
| 337 | 356 |
| 361 | 383 |
| 388 | 410 |
| 420 | 439 |
