You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004725_00090
You are here: Home > Sequence: MGYG000004725_00090
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA9475 sp900554075 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; UBA9475; UBA9475 sp900554075 | |||||||||||
| CAZyme ID | MGYG000004725_00090 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | Phosphoglycerol transferase I | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 256159; End: 259074 Strand: - | |||||||||||
Full Sequence Download help
| MLFNILNSFI FWAAWIAIPM VMEIIPSLGS LFILLKRRKS YQKKETPALY PEISIIIPVY | 60 |
| NSADTLEGCL RSIADSTYPN DAIRVFLVNN LSRDNSFEVF TQCQEMFPEL HMQWLNAQQG | 120 |
| KSRALNLALY NSSGKYIVHI DSDGFLEAHA LTNLVDLFES DLSCNCVTGS ILTIPEKIEE | 180 |
| YTGSGLLLRR MEFLEYAQAF LAGRNYASET NTIYTLSGAF SAFRKSAILK SWLYNTDTIC | 240 |
| EDTQLTFQMR YLQKEKVRIS VDSIFLTDPI EDLDKLYTQR QRWQRGSLEV SRMFMGTDEL | 300 |
| KARKLFGDVN VKTLMYDHTF AFPRMIWYLA TICMLFLGFS GKTILMATAL LFGLYTLCGY | 360 |
| FYFFSSLAFL EKFPDLRSYY KKQWWLVPLL PFFNLMVFFI RMAGIINSIG TDSSWKTRTF | 420 |
| TDERKSFAQI IRQDFSKVSG WMGKIRSRVN GDVDKMYAPK TLDQRLHGSV WAYIGVFAAF | 480 |
| AAALVLTVMV AWGTSAFSTP FESLVSTFFS STQGTASEVY LNGLKACLPP ILVALAAAVG | 540 |
| LIVLDRRQTK RVLSWSESPR RQRAAGQILR RRNTYAVLTC LFLAGSLIFA NAAYGVSDYV | 600 |
| GAKLVNSTLY ETHYADPRSV AITPPDQRPN LLYIYMESME TTYGSIDQGG ALEQSLIPNL | 660 |
| TRLARENVSF SQTDKPLGGF YSGTGTSWTM AALYATSSGA HFALPAGGSQ SAQSDHFAAG | 720 |
| LWTLGDVLAQ NGYQNEFLLG SSAEFSGLDA FFLQHGDYAL FDYPAAIDKG LLPQNYHQWW | 780 |
| GFEDAKLYEY AKAELTRLSQ SGQPFNLTMV TADTHFPEGY ICERCPTDKG ERAANVVACA | 840 |
| DAQLGEFIDW CKAQPFFENT VIVITGDHPR MDNVVVGDTP WDQRGVYDCF INARADRKGD | 900 |
| LRNRRFTAMD IYPTTLAALG FTIDGDTLGL GVDLFSGRET LAERLGYEAL DQELAKNDPY | 960 |
| YIPNLAPELL D | 971 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 54 | 228 | 1.3e-22 | 0.9705882352941176 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR03111 | glyc2_xrt_Gpos1 | 0.0 | 4 | 438 | 2 | 437 | putative glycosyltransferase, exosortase G-associated. Members of this protein family are probable glycosyltransferases of family 2, whose genes are near those for the exosortase homolog XrtG (TIGR03110), which is restricted to Gram-positive bacteria. Other genes in the conserved gene neighborhood include a 6-pyruvoyl tetrahydropterin synthase homolog (TIGR03112) and an uncharacterized intergral membrane protein (TIGR03766). |
| COG1215 | BcsA | 3.77e-55 | 1 | 426 | 1 | 430 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| cd16015 | LTA_synthase | 1.19e-53 | 629 | 920 | 1 | 283 | Lipoteichoic acid synthase like. Lipoteichoic acid (LTA) is an important cell wall polymer found in Gram-positive bacteria. It may contain long chains of ribitol or glycerol phosphate. LTA synthase catalyzes the reaction to extend the polymer by the repeated addition of glycerolphosphate (GroP) subunits to the end of the growing chain. |
| PRK03776 | PRK03776 | 2.32e-44 | 564 | 943 | 93 | 465 | phosphatidylglycerol--membrane-oligosaccharide glycerophosphotransferase. |
| PRK12363 | PRK12363 | 8.62e-44 | 619 | 936 | 147 | 457 | phosphoglycerol transferase I; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACR73592.1 | 4.63e-206 | 5 | 453 | 5 | 454 |
| QNU66959.1 | 1.22e-160 | 7 | 452 | 9 | 454 |
| QOX63067.1 | 3.50e-153 | 7 | 454 | 9 | 453 |
| CBK84035.1 | 6.93e-153 | 7 | 456 | 6 | 456 |
| ALU36827.1 | 4.58e-152 | 7 | 450 | 13 | 457 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3LXQ_A | 4.78e-15 | 608 | 934 | 69 | 367 | TheCrystal Structure of a Protein in the Alkaline Phosphatase Superfamily from Vibrio parahaemolyticus to 1.95A [Vibrio parahaemolyticus],3LXQ_B The Crystal Structure of a Protein in the Alkaline Phosphatase Superfamily from Vibrio parahaemolyticus to 1.95A [Vibrio parahaemolyticus] |
| 6YV7_B | 4.26e-10 | 38 | 155 | 29 | 149 | MannosyltransferasePcManGT from Pyrobaculum calidifontis [Pyrobaculum calidifontis JCM 11548],6YV8_B Mannosyltransferase PcManGT from Pyrobaculum calidifontis in complex with GDP and Mn2+ [Pyrobaculum calidifontis JCM 11548],6YV9_A Mannosyltransferase PcManGT from Pyrobaculum calidifontis in complex with GDP-Man and Mn2+ [Pyrobaculum calidifontis JCM 11548] |
| 6YV7_A | 4.28e-10 | 38 | 155 | 30 | 150 | MannosyltransferasePcManGT from Pyrobaculum calidifontis [Pyrobaculum calidifontis JCM 11548],6YV8_A Mannosyltransferase PcManGT from Pyrobaculum calidifontis in complex with GDP and Mn2+ [Pyrobaculum calidifontis JCM 11548],6YV9_B Mannosyltransferase PcManGT from Pyrobaculum calidifontis in complex with GDP-Man and Mn2+ [Pyrobaculum calidifontis JCM 11548] |
| 5HEA_A | 1.54e-09 | 53 | 154 | 7 | 105 | CgTstructure in hexamer [Streptococcus parasanguinis FW213],5HEA_B CgT structure in hexamer [Streptococcus parasanguinis FW213],5HEA_C CgT structure in hexamer [Streptococcus parasanguinis FW213],5HEC_A CgT structure in dimer [Streptococcus parasanguinis FW213],5HEC_B CgT structure in dimer [Streptococcus parasanguinis FW213] |
| 2Z86_A | 8.39e-07 | 41 | 167 | 360 | 488 | Crystalstructure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP [Escherichia coli],2Z86_B Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP [Escherichia coli],2Z86_C Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP [Escherichia coli],2Z86_D Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A8AM07 | 2.48e-35 | 564 | 938 | 93 | 461 | Phosphoglycerol transferase I OS=Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696) OX=290338 GN=mdoB PE=3 SV=1 |
| A9N7A9 | 3.28e-35 | 564 | 938 | 93 | 461 | Phosphoglycerol transferase I OS=Salmonella paratyphi B (strain ATCC BAA-1250 / SPB7) OX=1016998 GN=mdoB PE=3 SV=1 |
| B4T4E6 | 3.28e-35 | 564 | 938 | 93 | 461 | Phosphoglycerol transferase I OS=Salmonella newport (strain SL254) OX=423368 GN=mdoB PE=3 SV=1 |
| Q57G63 | 3.28e-35 | 564 | 938 | 93 | 461 | Phosphoglycerol transferase I OS=Salmonella choleraesuis (strain SC-B67) OX=321314 GN=mdoB PE=3 SV=1 |
| B4TU18 | 4.36e-35 | 564 | 938 | 93 | 461 | Phosphoglycerol transferase I OS=Salmonella schwarzengrund (strain CVM19633) OX=439843 GN=mdoB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
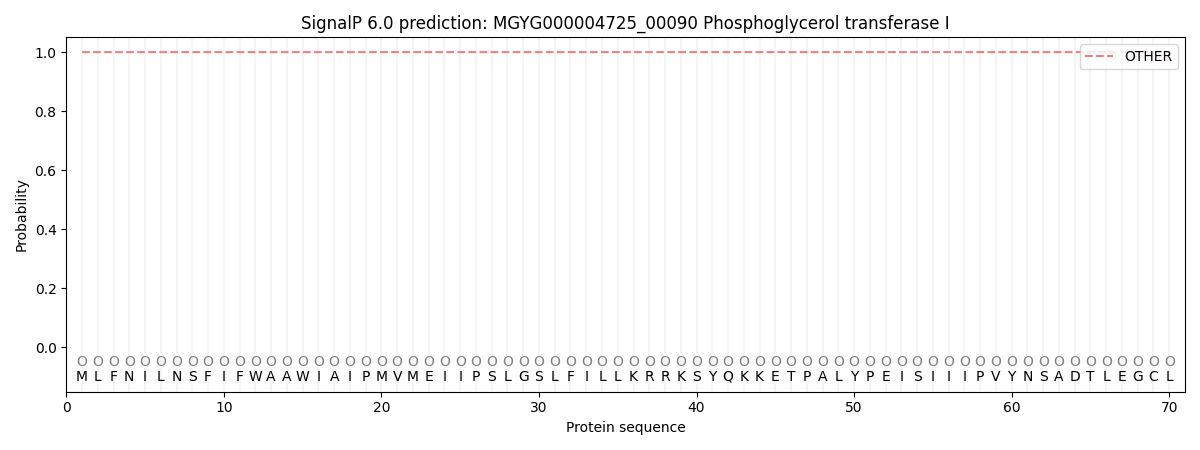
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999981 | 0.000050 | 0.000001 | 0.000000 | 0.000000 | 0.000001 |
TMHMM Annotations download full data without filtering help
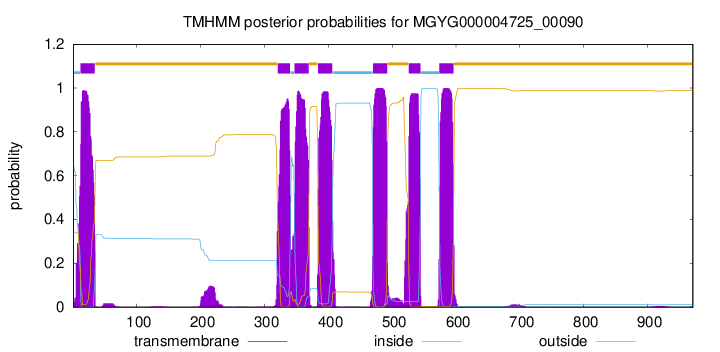
| start | end |
|---|---|
| 12 | 34 |
| 321 | 340 |
| 347 | 369 |
| 384 | 406 |
| 470 | 492 |
| 526 | 544 |
| 574 | 596 |
