You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000650_00104
You are here: Home > Sequence: MGYG000000650_00104
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Enterocloster; | |||||||||||
| CAZyme ID | MGYG000000650_00104 | |||||||||||
| CAZy Family | GT27 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 33518; End: 37420 Strand: - | |||||||||||
Full Sequence Download help
| MKDMSYEILD LFKAYGSGPD GIQRILEENS NPALLYALSP LRKNQLEWLD TTGEEEVLQI | 60 |
| GSDYGALTGL LASRCAHVTV MDEADENLEV NRQRNSDYVN ITYGFSFERG VENQGKLFDL | 120 |
| VLVSSLEPGR DIQEAVSFGA AFLKQGGRLI FSCENDLGLR FLAGGEHMEA SCTRQEAKEL | 180 |
| CEGLGFSRVE FYYPVTDQKL PSSVYSDRYL PSRGEVGAGA ALEGPRYACF QEEAVYDGME | 240 |
| GGGLFGSFAN GFVVIASNGG CETTCFAKYN RTRRKEFQTR TRIWEKEEGC QVEKSALSPE | 300 |
| GKSHLEAFGA HYESLSRCYP HICFLKPVMG ERKDRITFPF LKGRTLAECL GEQIRDGKAP | 360 |
| VCELEKALEA VIGRPEIPLK PFVPTPELEQ VFGNVPDLGG MSYEISNIDG LFENLMVQEE | 420 |
| NGEEKLYCLD YEWVFDFPIP VSFVRYRILA YFYYRYEGSL CYGSLEGFLK EFGIEGDTAE | 480 |
| LYANMEKAFQ SYVHGDALQG YLANYIHEVT DMEQIRSMKK ELEKARDRIN QLQEEVEERN | 540 |
| LFIRKEQELK RLTNNHVANL EIMIKDLRHE IDELGKLATY LNGHEALVFK ARRKLGVQVS | 600 |
| KAFPKGTRKR KALDYCVSTV KHPLRYGKMY ATKEGRRRIK GDFAIGEDYL KYGKLVFPQV | 660 |
| PKEGPMVSIV IPCYNQIGYT YACLQSILEF TKDVTYEVII ADDVSTDATA QLGQYTEGLV | 720 |
| ICRNQTNQGF LRNCNQAARA ARGKYIMFLN NDTKVTEGWL SSLVNLIQSD ASIGMVGSKL | 780 |
| VYPDGRLQEA GGIIWSDGSG WNYGRLDDPD KPEYNYVKDV DYISGAAILL STKLWKQIGG | 840 |
| FDERFAPAYC EDSDLAFEVR KAGYRVVYQP LSKVIHFEGI SNGTDVNGTG LKRYQVENSR | 900 |
| KLKEKWAEEL KKQCVNTGNP NPFRARERSQ GKQVILVVDH YVPTFDKDAG SKTTYQYLKM | 960 |
| FLKKGYVVKF LGDNFLHEEP YTTILEQMGI EVLYGEAMSA GIWDWLKKNG DEIDFAYLNR | 1020 |
| PHIATKYVDF IREHTNMKVI YYGHDLHFLR LGREYKLTKD FAVKREADYW KSVELTMMHK | 1080 |
| ADISYYPSYV EIDAIHEMDA SIPAKAITAY VYDQFLDHIE EDFAKREGLL FVGGFAHPPN | 1140 |
| ADAVLWFAEE IFPLIRRALP EANFYIVGSK VTEEIQALEK PGNGIVVKGF VTEEELAELY | 1200 |
| STCRMVVVPL RYGAGVKGKV VEAIYNGAPI VTTSTGAEGI PSGETVLEIE DEAAAFAAKT | 1260 |
| ISLYQDPERL AQMCRRTQEY IREYYSLDGA WNVIKDDFAR | 1300 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT27 | 668 | 917 | 1.6e-24 | 0.8813559322033898 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd04186 | GT_2_like_c | 7.00e-46 | 669 | 878 | 1 | 166 | Subfamily of Glycosyltransferase Family GT2 of unknown function. GT-2 includes diverse families of glycosyltransferases with a common GT-A type structural fold, which has two tightly associated beta/alpha/beta domains that tend to form a continuous central sheet of at least eight beta-strands. These are enzymes that catalyze the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. Glycosyltransferases have been classified into more than 90 distinct sequence based families. |
| COG1216 | GT2 | 1.62e-45 | 665 | 883 | 3 | 226 | Glycosyltransferase, GT2 family [Carbohydrate transport and metabolism]. |
| pfam13692 | Glyco_trans_1_4 | 1.53e-24 | 1129 | 1266 | 4 | 138 | Glycosyl transferases group 1. |
| COG1215 | BcsA | 1.85e-22 | 660 | 882 | 49 | 269 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| pfam00535 | Glycos_transf_2 | 2.05e-22 | 668 | 788 | 1 | 124 | Glycosyl transferase family 2. Diverse family, transferring sugar from UDP-glucose, UDP-N-acetyl- galactosamine, GDP-mannose or CDP-abequose, to a range of substrates including cellulose, dolichol phosphate and teichoic acids. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASN93838.1 | 0.0 | 1 | 1298 | 1 | 1313 |
| QRP41495.1 | 0.0 | 1 | 1298 | 1 | 1313 |
| QJU21849.1 | 0.0 | 1 | 1298 | 1 | 1313 |
| ANU46567.1 | 0.0 | 1 | 1298 | 1 | 1314 |
| QQQ98718.1 | 0.0 | 1 | 1298 | 1 | 1314 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5HEA_A | 3.96e-07 | 663 | 772 | 3 | 114 | CgTstructure in hexamer [Streptococcus parasanguinis FW213],5HEA_B CgT structure in hexamer [Streptococcus parasanguinis FW213],5HEA_C CgT structure in hexamer [Streptococcus parasanguinis FW213],5HEC_A CgT structure in dimer [Streptococcus parasanguinis FW213],5HEC_B CgT structure in dimer [Streptococcus parasanguinis FW213] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P55465 | 4.29e-21 | 628 | 906 | 581 | 874 | Uncharacterized protein y4gI OS=Sinorhizobium fredii (strain NBRC 101917 / NGR234) OX=394 GN=NGR_a03550 PE=4 SV=1 |
| Q50864 | 1.89e-17 | 665 | 885 | 574 | 801 | O-antigen biosynthesis protein RfbC OS=Myxococcus xanthus OX=34 GN=rfbC PE=4 SV=1 |
| Q8MYY6 | 4.52e-11 | 667 | 911 | 112 | 381 | Putative polypeptide N-acetylgalactosaminyltransferase 13 OS=Drosophila melanogaster OX=7227 GN=pgant13 PE=2 SV=2 |
| P9WLV8 | 3.39e-10 | 665 | 806 | 17 | 148 | Uncharacterized protein MT1568 OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=MT1568 PE=4 SV=1 |
| P9WLV9 | 3.39e-10 | 665 | 806 | 17 | 148 | Uncharacterized protein Rv1518 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=Rv1518 PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
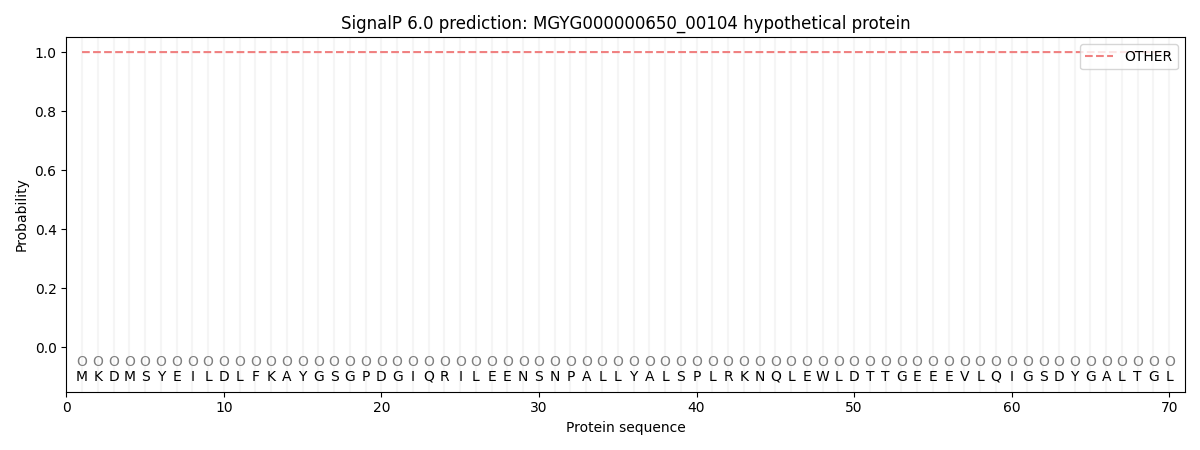
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000054 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
