You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001626_00593
You are here: Home > Sequence: MGYG000001626_00593
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
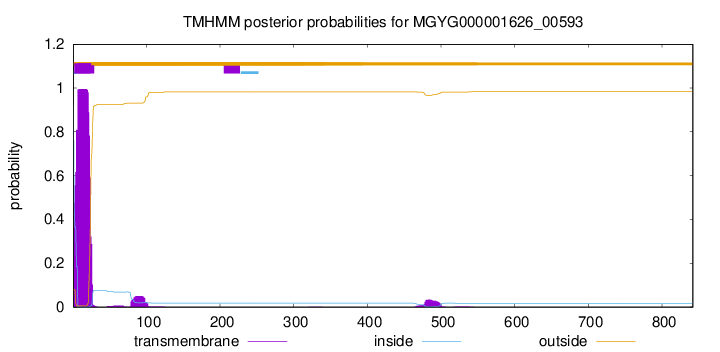
TMHMM annotations
Basic Information help
| Species | Phascolarctobacterium sp900544795 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Acidaminococcales; Acidaminococcaceae; Phascolarctobacterium; Phascolarctobacterium sp900544795 | |||||||||||
| CAZyme ID | MGYG000001626_00593 | |||||||||||
| CAZy Family | GT30 | |||||||||||
| CAZyme Description | Tetraacyldisaccharide 4'-kinase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 65174; End: 67702 Strand: - | |||||||||||
Full Sequence Download help
| MYIIYNFLVL CVCCLFVLPY FLYRCICEAG FTTRMRQSLG GLRDEEIAAV ALKDCIWIHG | 60 |
| ASVGEIVATS PLVKEIRKAY PNAPILVSAV TVGGYNMAKQ IIREADTIIY FPLDMPFVSE | 120 |
| SFVKRIQPRI FMPVETELWP NFLRAIRERN IPVMMVNGRI SEKSVKTYRY LYGIWDDMLH | 180 |
| TVSRFCMQSS IDADYISHLG ADPRKIYVTG NTKFDQTYAE VTPEDLENYK KELGIESAYP | 240 |
| VIVAGSTHPT EEKVLFDVFK DIIKTYPDAR LVIAPRKTGR ADEISKLAKQ YHWDTGLRSK | 300 |
| LLEMEGERPK YPVLLIDTIG ELGRIYSVGD IVFVGGSLIK HGGHNVLEPA AHAKPILVGP | 360 |
| NMQNFKDSYA LLSKVGACKM VNSSAELTKE VLEIAGNDAR RQQMGAASLQ VIKENRGAAV | 420 |
| RSIEYLQDLL TLTTVDSKVE ASYPTNTRNI HDEGGGRLRH GDAVIQYLYQ LAHGPNTPFF | 480 |
| GWILLGILRM FSYVYEFGVC CKLNLYKSGL LKQKKLDCCV ISIGNITVGG TGKTPTAQKV | 540 |
| AVIIKNMGYR VVILNRGYRS HWDQEMGVVS DGKKIFMTAY EAGDEAYLMA KTLPGVPVII | 600 |
| GKNRALTGQF AVDKMNAEVI IMDDGYQHWH LHRDLDVVLV DTFNMFGNGC LLPRGTLREP | 660 |
| LTHLDRAGLF LLTKTDQSSQ FSRQELRDTL DKYNGQAPII ESVHHPKNFV EIADWYKGIH | 720 |
| ENAKDLSELQ GKKVMVFSAI GNPSSFEQTL STIGIEIIEA IRYPDHHDYG MLEMQYISER | 780 |
| AINKEVVAMV TTGKDAVKIP TEFIYFNREV PLYILNMDIM ITEGREEFEK TILGAIQKET | 840 |
| KQ | 842 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT30 | 53 | 216 | 1e-55 | 0.9209039548022598 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK05749 | PRK05749 | 1.43e-122 | 1 | 430 | 2 | 421 | 3-deoxy-D-manno-octulosonic-acid transferase; Reviewed |
| COG1519 | KdtA | 1.22e-113 | 7 | 430 | 5 | 419 | 3-deoxy-D-manno-octulosonic-acid transferase [Cell wall/membrane/envelope biogenesis]. |
| pfam02606 | LpxK | 6.41e-85 | 487 | 837 | 4 | 318 | Tetraacyldisaccharide-1-P 4'-kinase. This family consists of tetraacyldisaccharide-1-P 4'-kinase also known as Lipid-A 4'-kinase or Lipid A biosynthesis protein LpxK, EC:2.7.1.130. This enzyme catalyzes the reaction: ATP + 2,3-bis(3-hydroxytetradecanoyl)-D -glucosaminyl-(beta-D-1,6)-2,3-bis(3-hydroxytetradecanoyl)-D-glu cosam inyl beta-phosphate <=> ADP + 2,3,2',3'-tetrakis(3-hydroxytetradecanoyl)-D- glucosaminyl-1,6-beta-D-glucosamine 1,4'-bisphosphate. This enzyme is involved in the synthesis of lipid A portion of the bacterial lipopolysaccharide layer (LPS). The family contains a P-loop motif at the N-terminus. |
| PRK00652 | lpxK | 2.00e-68 | 481 | 838 | 12 | 325 | tetraacyldisaccharide 4'-kinase; Reviewed |
| pfam04413 | Glycos_transf_N | 3.55e-68 | 53 | 216 | 13 | 176 | 3-Deoxy-D-manno-octulosonic-acid transferase (kdotransferase). Members of this family transfer activated sugars to a variety of substrates, including glycogen, fructose-6-phosphate and lipopolysaccharides. Members of the family transfer UDP, ADP, GDP or CMP linked sugars. The Glycos_transf_N region is flanked at the N-terminus by a signal peptide and at the C-terminus by Glycos_transf_1 (pfam00534). The eukaryotic glycogen synthases may be distant members of this bacterial family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBG63225.1 | 0.0 | 1 | 840 | 1 | 840 |
| QNP77900.1 | 0.0 | 1 | 840 | 1 | 840 |
| QTV78313.1 | 0.0 | 1 | 839 | 1 | 839 |
| AEQ21866.1 | 0.0 | 1 | 841 | 1 | 841 |
| ADB48040.1 | 0.0 | 1 | 839 | 1 | 839 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4EHX_A | 4.40e-42 | 488 | 832 | 6 | 310 | Crystalstructure of LpxK from Aquifex aeolicus at 1.9 angstrom resolution [Aquifex aeolicus VF5],4EHY_A Crystal structure of LpxK from Aquifex aeolicus in complex with ADP/Mg2+ at 2.2 angstrom resolution [Aquifex aeolicus VF5],4ITL_A Crystal structure of LpxK from Aquifex aeolicus in complex with AMP-PCP at 2.1 angstrom resolution [Aquifex aeolicus VF5],4ITM_A Crystal structure of 'apo' form LpxK from Aquifex aeolicus in complex with ATP at 2.2 angstrom resolution [Aquifex aeolicus VF5],4ITN_A Crystal structure of 'compact P-loop' LpxK from Aquifex aeolicus in complex with chloride at 2.2 angstrom resolution [Aquifex aeolicus VF5],4LKV_A Determinants of lipid substrate and membrane binding for the tetraacyldisaccharide-1-phosphate 4 -kinase LpxK [Aquifex aeolicus VF5],4LKV_B Determinants of lipid substrate and membrane binding for the tetraacyldisaccharide-1-phosphate 4 -kinase LpxK [Aquifex aeolicus VF5],4LKV_C Determinants of lipid substrate and membrane binding for the tetraacyldisaccharide-1-phosphate 4 -kinase LpxK [Aquifex aeolicus VF5],4LKV_D Determinants of lipid substrate and membrane binding for the tetraacyldisaccharide-1-phosphate 4 -kinase LpxK [Aquifex aeolicus VF5] |
| 4EHW_A | 4.62e-42 | 488 | 832 | 8 | 312 | Crystalstructure of LpxK from Aquifex aeolicus at 2.3 angstrom resolution [Aquifex aeolicus VF5] |
| 2XCI_A | 1.54e-33 | 51 | 394 | 35 | 343 | Membrane-embeddedmonofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCI_B Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCI_C Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCI_D Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCU_A Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus],2XCU_B Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus],2XCU_C Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus],2XCU_D Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus] |
| 4BFC_A | 2.63e-13 | 241 | 423 | 44 | 224 | ChainA, 3-DEOXY-D-MANNO-OCTULOSONIC-ACID TRANSFERASE [Acinetobacter baumannii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8CZC3 | 1.60e-70 | 467 | 821 | 12 | 371 | Tetraacyldisaccharide 4'-kinase OS=Halothermothrix orenii (strain H 168 / OCM 544 / DSM 9562) OX=373903 GN=lpxK PE=3 SV=1 |
| Q2LVL1 | 6.15e-69 | 487 | 833 | 26 | 368 | Tetraacyldisaccharide 4'-kinase OS=Syntrophus aciditrophicus (strain SB) OX=56780 GN=lpxK PE=3 SV=1 |
| P44806 | 8.22e-62 | 56 | 430 | 53 | 423 | 3-deoxy-D-manno-octulosonic acid transferase OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=waaA PE=1 SV=1 |
| B8F928 | 3.12e-61 | 486 | 837 | 25 | 365 | Tetraacyldisaccharide 4'-kinase OS=Desulfatibacillum aliphaticivorans OX=218208 GN=lpxK PE=3 SV=1 |
| A7HDI5 | 1.63e-59 | 483 | 838 | 19 | 362 | Tetraacyldisaccharide 4'-kinase OS=Anaeromyxobacter sp. (strain Fw109-5) OX=404589 GN=lpxK PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
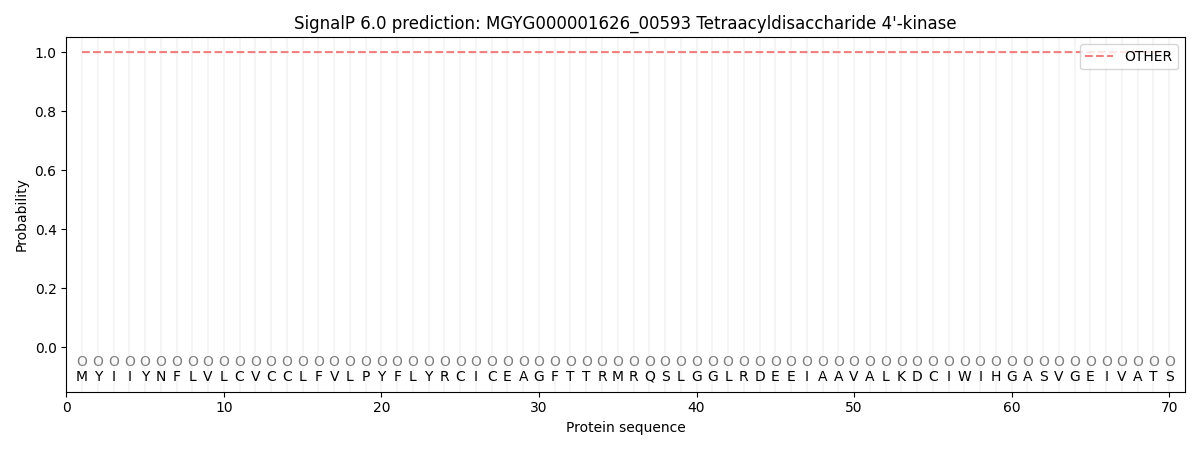
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000019 | 0.000016 | 0.000001 | 0.000000 | 0.000000 | 0.000000 |