You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004685_00134
You are here: Home > Sequence: MGYG000004685_00134
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-245 sp900758165 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; TANB77; CAG-508; CAG-245; CAG-245 sp900758165 | |||||||||||
| CAZyme ID | MGYG000004685_00134 | |||||||||||
| CAZy Family | GT32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17148; End: 18968 Strand: + | |||||||||||
Full Sequence Download help
| MEKIIHYCWF GDKPLSKLAK KCIKSWKKFL PDYKIIKWTE ENVNLNECIF IKEAYANKKW | 60 |
| AFVADYVRTK ALNEYGGIYF DTDMEVTKDI SDLLESNKTF LGVEDTGYVA VGVWYEREKK | 120 |
| SFLSSELLKR YRSIKKFDIN KMADMSIPKI VSEILEPYGL KKGGTKIQLL ENDIKIYPRD | 180 |
| YFYPYSFHRD NNVFSENTCM IHYYDASWLP KRDKREMMLV RRFGRDKTNK ILNTYRRLKQ | 240 |
| LIIKCLKIPL FPIVLYRHHK AKMIVITDSY LNRLNETINC IEKNKDKEYI TIYNKLWLGV | 300 |
| TSATNELFEN LVDCNEIYRK KDVKAIGNAI LNANIKQLVF SSFALGWEEL VKYLRTKNKN | 360 |
| IKIKVFWHGS HSQILDPYGW KRNLEFIHLH QSGYVDAIGT CKESLMNFYK KQGYNAYFIT | 420 |
| NKVSEIKITK EESKIKTDKD KKVIGIYAAK CDDWRKNMYS QMAAVKLIEN AVIDMVPLNE | 480 |
| QAKTFAEQYG IEITGVEKPI SREKMMQRMA KNDINLYVTF SECAPMLPLE SMELGVPCIT | 540 |
| GNNHHYFKNN KLEKYIVINN ETDIEEIKRK ILECIENKAK IMQYYKQFSE KNIEESKEEV | 600 |
| KQFLNA | 606 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT32 | 18 | 96 | 3.1e-22 | 0.9666666666666667 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3774 | OCH1 | 1.36e-19 | 2 | 214 | 83 | 285 | Mannosyltransferase OCH1 or related enzyme [Cell wall/membrane/envelope biogenesis]. |
| pfam04488 | Gly_transf_sug | 4.55e-13 | 20 | 94 | 5 | 87 | Glycosyltransferase sugar-binding region containing DXD motif. The DXD motif is a short conserved motif found in many families of glycosyltransferases, which add a range of different sugars to other sugars, phosphates and proteins. DXD-containing glycosyltransferases all use nucleoside diphosphate sugars as donors and require divalent cations, usually manganese. The DXD motif is expected to play a carbohydrate binding role in sugar-nucleoside diphosphate and manganese dependent glycosyltransferases. |
| pfam05704 | Caps_synth | 4.26e-10 | 1 | 95 | 46 | 145 | Capsular polysaccharide synthesis protein. This family consists of several capsular polysaccharide proteins. Capsular polysaccharide (CPS) is a major virulence factor in Streptococcus pneumoniae. |
| COG0438 | RfaB | 7.99e-06 | 338 | 599 | 87 | 360 | Glycosyltransferase involved in cell wall bisynthesis [Cell wall/membrane/envelope biogenesis]. |
| pfam00534 | Glycos_transf_1 | 1.93e-04 | 472 | 580 | 38 | 147 | Glycosyl transferases group 1. Mutations in this domain of PIGA lead to disease (Paroxysmal Nocturnal haemoglobinuria). Members of this family transfer activated sugars to a variety of substrates, including glycogen, Fructose-6-phosphate and lipopolysaccharides. Members of this family transfer UDP, ADP, GDP or CMP linked sugars. The eukaryotic glycogen synthases may be distant members of this family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CAI33206.1 | 1.39e-53 | 1 | 231 | 2 | 231 |
| ASO97438.1 | 5.42e-52 | 1 | 231 | 2 | 231 |
| AUC46774.1 | 5.42e-52 | 1 | 231 | 2 | 231 |
| QCO91874.1 | 5.42e-52 | 1 | 231 | 2 | 231 |
| ASO97422.1 | 5.42e-52 | 1 | 231 | 2 | 231 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
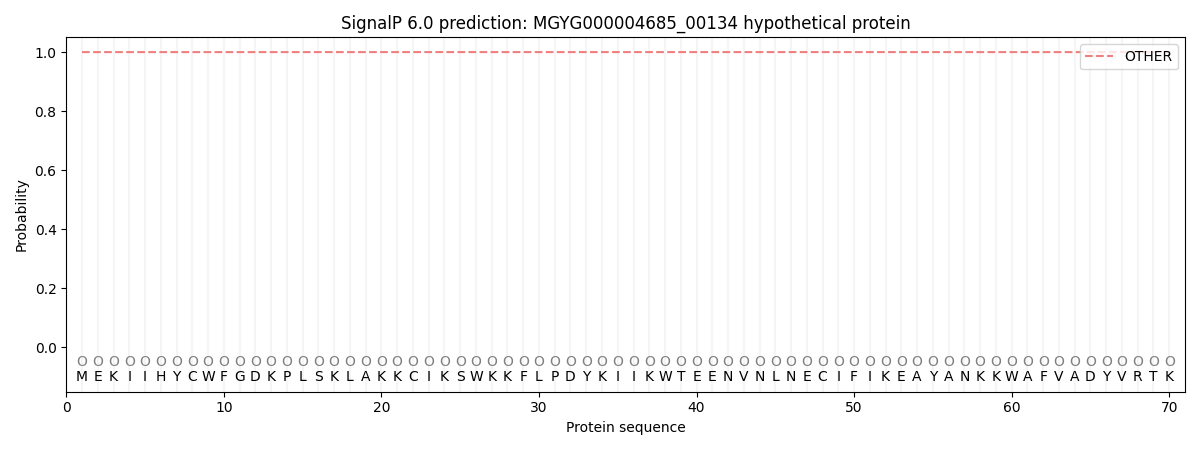
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000034 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
