You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003949_00116
You are here: Home > Sequence: MGYG000003949_00116
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Eubacterium_F sp900539115 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_F; Eubacterium_F sp900539115 | |||||||||||
| CAZyme ID | MGYG000003949_00116 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 132562; End: 135744 Strand: + | |||||||||||
Full Sequence Download help
| MYQDILTIAV VTVFAFFAIY WMWFQGEVDG SRLIAKNSMK QQKFNIILVL VAAFIVKAIL | 60 |
| AVKNEGYSVD ITCFRSWSET VFTGGLSHFY EGDVTDYPPG YMLVLWLVAA IRHLFNIDNY | 120 |
| STTGLFMIKL FPMLFDLAAA YLIYVMAKKR FSEGMSLMLS TLYALNPAVV IDSSVWGQVD | 180 |
| GVLCCMVLLT CYLFMEEKRI LAYFAFCAGV LLKPQMVMFA PIIIYSIIEQ VFLHDFNTKK | 240 |
| MLRDLIGGLA SLAAMVLVAL PFGLEKSLEK YTGTLGMFEY CSVNAYNFWA IVGKNWHEQT | 300 |
| EKFLFIPAQT WGTIAIIAAV VLSAVVFFKM KNDKSKYFLS MAVVISVMFC FSVRMHERYL | 360 |
| FPAVALMLAA FIYRPCKELF YGFIGYTLVV YMNTAHVYYT AVELGTTGPT GYIIGAVAVL | 420 |
| TLFLFGYLLY TIFKKSEIVQ EANVDEGGKS KKKRQTVQKQ QKKRFEWKII PTVVMEKMTK | 480 |
| TDYIILFAIM LVYGIFAFHD LGKTSAPETG WEGTQAGQQI VLDFGEVKNL KEFEVFSGSY | 540 |
| ENRSLYMELS NDGTNYVSGG TAKVSDVFKW NNVQLVADGG SEATTAFNVT TRYIRFTTVE | 600 |
| DLVTLKEMVF KDADGNLVVP VNKDQYPELF DEQDEYDDAH TFRSGTYFDE IYHARTAYEM | 660 |
| IHDLYNYENT HPPLGKIFIS LGIRIFGMNP FGWRIIGTLF GIGMLPFLYL FGKRLFHQTW | 720 |
| VAGVVTTLFA FDFMHFTQTR IATIDVYGTF FIMAMFYFML RYAQTSFYDT EFKKTLIPLF | 780 |
| LSGLMMGLGC ASKWTAVYAA AGLAVFFAAI MFYRYMEYRR ACNNPGGSTG TIAHRHVIDV | 840 |
| FKSNFLKTIG ACVIFFIVIP GLIYLCSYIP FNDGTTDGLF TRMINNQKSM YSYHSQLEAT | 900 |
| HPYSSTWYEW PTMIRPVFYY CNTVANDMRE GISAFGNPLV WWAGIFAFLY MIYLVVKKAD | 960 |
| KTALFLVFAY LVQYVPWMLV PRCTFAYHYF PSVPFITLMI GYTLYRFIGD NKKKRIFAYV | 1020 |
| YAAAAVGLFL LFYPVLSGQP VSLEFVENGL RWLTGWVLVL | 1060 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 646 | 871 | 7.9e-55 | 0.905829596412556 |
| GT83 | 103 | 424 | 3.1e-18 | 0.5351851851851852 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16192 | PMT_4TMC | 1.96e-26 | 887 | 1053 | 7 | 197 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| COG5542 | COG5542 | 5.27e-17 | 44 | 375 | 41 | 364 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
| COG1928 | PMT1 | 1.70e-16 | 668 | 910 | 87 | 302 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| COG1928 | PMT1 | 1.42e-14 | 880 | 1058 | 498 | 695 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| pfam13231 | PMT_2 | 2.01e-13 | 671 | 807 | 2 | 129 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCN30459.1 | 5.14e-311 | 22 | 1059 | 18 | 1059 |
| QNM01664.1 | 1.45e-281 | 52 | 1059 | 42 | 1086 |
| BBF42044.1 | 8.72e-276 | 142 | 1059 | 2 | 921 |
| AIQ66842.1 | 2.36e-214 | 52 | 1057 | 18 | 1030 |
| ANY70643.1 | 9.87e-213 | 31 | 1058 | 4 | 1050 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WN04 | 5.96e-24 | 648 | 1056 | 73 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=pmt PE=3 SV=2 |
| P9WN05 | 5.96e-24 | 648 | 1056 | 73 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=pmt PE=1 SV=2 |
| L8F4Z2 | 1.79e-23 | 648 | 1056 | 67 | 515 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycolicibacterium smegmatis (strain MKD8) OX=1214915 GN=pmt PE=3 SV=1 |
| Q8NRZ6 | 1.79e-21 | 644 | 1056 | 59 | 519 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=pmt PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000021 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
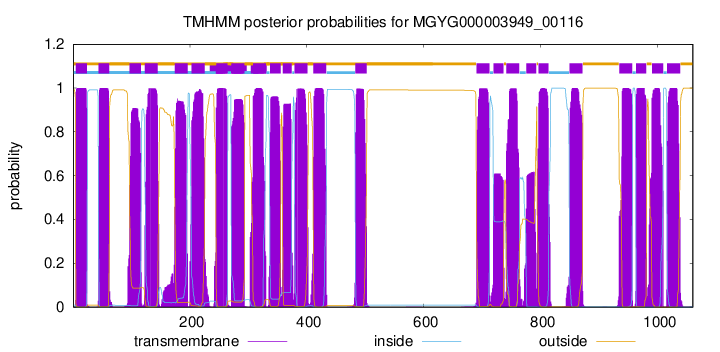
| start | end |
|---|---|
| 5 | 24 |
| 44 | 62 |
| 97 | 116 |
| 123 | 145 |
| 174 | 196 |
| 203 | 225 |
| 245 | 264 |
| 271 | 293 |
| 308 | 330 |
| 337 | 355 |
| 359 | 374 |
| 379 | 401 |
| 411 | 433 |
| 483 | 502 |
| 690 | 712 |
| 719 | 736 |
| 741 | 763 |
| 775 | 792 |
| 796 | 813 |
| 849 | 871 |
| 934 | 956 |
| 963 | 980 |
| 990 | 1009 |
| 1016 | 1038 |
