You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004887_00153
You are here: Home > Sequence: MGYG000004887_00153
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1417 sp002305575 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; UBA1417; UBA1417 sp002305575 | |||||||||||
| CAZyme ID | MGYG000004887_00153 | |||||||||||
| CAZy Family | GT4 | |||||||||||
| CAZyme Description | Glycosyltransferase Gtf1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 15111; End: 17552 Strand: - | |||||||||||
Full Sequence Download help
| MIKKMFSQLR KMKWRELRNH YWPRLRYPTY YRKLPIDDHA ILLEASHGKQ MQGNIYYLLK | 60 |
| VLASDPRYRE YRIYLSAHKT VAESFREALD RDGLNRVELA VVFTKKYMRL IASAKYLIND | 120 |
| TTFLPFFVKR PEQVYLNTWH GTPLKTLGKK VNNDLHNIGN VQKNFFMADY LLYPNVYTMK | 180 |
| HMVEDYMLED LSKAKILLAG YPRNTAFFDK KHAAEIREEL ELADKRVYAF MPTWRGVVGS | 240 |
| VDRKASTYIQ YYLYELDKRL HEDEVLYVNL HPLARKDVSF AGMAHIQPFP EAYETYAFLN | 300 |
| AADCLITDYS SVFYDFAVSR KKCVLFTYDE AEYFADRGLY RPLSALPFPS VKTVDELVEA | 360 |
| VRAPKSYDDT AFLREYCAYD NAEAAAQLCA RVILNEPSSA IEERDMPSNG KRNVLLYVGN | 420 |
| LAQNGITTSV MNLLRNVDRT RYNFYFTFES GKVRANRAIL QHLPSGTRYI ATTGKINLTF | 480 |
| WQKVVYLLFM ARLFPLAPYL RVMRTAYPME IRRNYGDAVF SDVIQFNGYE TKKILFFAAF | 540 |
| DANRVIYVHS DMQMEIRTRG NQRKNVLRYA YETYDHVAVV TEDIVEPTRS FLKKDRPFDL | 600 |
| GRNLIADRTI RKKGEKPIAF DPQTRCNVPF ETLMDIVARD AKRIVSVGRF SPEKGQRRLI | 660 |
| DAFVRVWQEN PASYLIIIGG NQRDGLYGQL CDYVSALPCR DHIVLILTMS NPFPLIKACD | 720 |
| GFILSSFYEG FGLVLAEADI LGLPVVSTDI TGPRRFMQAH GGVLVENSEA GLEDGLRRLL | 780 |
| NGQIPKLHID YDAYNREAVE EFYKLLDKDG KTK | 813 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam04464 | Glyphos_transf | 4.89e-55 | 35 | 394 | 1 | 360 | CDP-Glycerol:Poly(glycerophosphate) glycerophosphotransferase. Wall-associated teichoic acids are a heterogeneous class of phosphate-rich polymers that are covalently linked to the cell wall peptidoglycan of gram-positive bacteria. They consist of a main chain of phosphodiester-linked polyols and/or sugar moieties attached to peptidoglycan via a linkage unit. CDP-glycerol:poly(glycerophosphate) glycerophosphotransferase is responsible for the polymerization of the main chain of the teichoic acid by sequential transfer of glycerol-phosphate units from CDP-glycerol to the linkage unit lipid. |
| cd03811 | GT4_GT28_WabH-like | 2.85e-41 | 413 | 803 | 1 | 349 | family 4 and family 28 glycosyltransferases similar to Klebsiella WabH. This family is most closely related to the GT1 family of glycosyltransferases. WabH in Klebsiella pneumoniae has been shown to transfer a GlcNAc residue from UDP-GlcNAc onto the acceptor GalUA residue in the cellular outer core. |
| COG1887 | TagB | 2.18e-33 | 31 | 381 | 19 | 366 | CDP-glycerol glycerophosphotransferase, TagB/SpsB family [Cell wall/membrane/envelope biogenesis, Lipid transport and metabolism]. |
| pfam00534 | Glycos_transf_1 | 3.13e-24 | 640 | 796 | 1 | 158 | Glycosyl transferases group 1. Mutations in this domain of PIGA lead to disease (Paroxysmal Nocturnal haemoglobinuria). Members of this family transfer activated sugars to a variety of substrates, including glycogen, Fructose-6-phosphate and lipopolysaccharides. Members of this family transfer UDP, ADP, GDP or CMP linked sugars. The eukaryotic glycogen synthases may be distant members of this family. |
| cd03801 | GT4_PimA-like | 1.50e-23 | 495 | 781 | 56 | 332 | phosphatidyl-myo-inositol mannosyltransferase. This family is most closely related to the GT4 family of glycosyltransferases and named after PimA in Propionibacterium freudenreichii, which is involved in the biosynthesis of phosphatidyl-myo-inositol mannosides (PIM) which are early precursors in the biosynthesis of lipomannans (LM) and lipoarabinomannans (LAM), and catalyzes the addition of a mannosyl residue from GDP-D-mannose (GDP-Man) to the position 2 of the carrier lipid phosphatidyl-myo-inositol (PI) to generate a phosphatidyl-myo-inositol bearing an alpha-1,2-linked mannose residue (PIM1). Glycosyltransferases catalyze the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. The acceptor molecule can be a lipid, a protein, a heterocyclic compound, or another carbohydrate residue. This group of glycosyltransferases is most closely related to the previously defined glycosyltransferase family 1 (GT1). The members of this family may transfer UDP, ADP, GDP, or CMP linked sugars. The diverse enzymatic activities among members of this family reflect a wide range of biological functions. The protein structure available for this family has the GTB topology, one of the two protein topologies observed for nucleotide-sugar-dependent glycosyltransferases. GTB proteins have distinct N- and C- terminal domains each containing a typical Rossmann fold. The two domains have high structural homology despite minimal sequence homology. The large cleft that separates the two domains includes the catalytic center and permits a high degree of flexibility. The members of this family are found mainly in certain bacteria and archaea. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL33357.1 | 1.43e-275 | 15 | 807 | 13 | 808 |
| CBK97012.1 | 1.16e-274 | 15 | 807 | 13 | 808 |
| QQQ92281.1 | 2.32e-204 | 27 | 808 | 20 | 798 |
| ANU74732.2 | 3.62e-204 | 27 | 808 | 34 | 812 |
| ASU27537.1 | 3.62e-204 | 27 | 808 | 34 | 812 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3L7I_A | 4.95e-23 | 36 | 385 | 350 | 707 | Structureof the Wall Teichoic Acid Polymerase TagF [Staphylococcus epidermidis RP62A],3L7I_B Structure of the Wall Teichoic Acid Polymerase TagF [Staphylococcus epidermidis RP62A],3L7I_C Structure of the Wall Teichoic Acid Polymerase TagF [Staphylococcus epidermidis RP62A],3L7I_D Structure of the Wall Teichoic Acid Polymerase TagF [Staphylococcus epidermidis RP62A] |
| 3L7J_A | 3.47e-22 | 36 | 385 | 350 | 707 | ChainA, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A],3L7J_B Chain B, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A],3L7J_C Chain C, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A],3L7J_D Chain D, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A],3L7K_A Chain A, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A],3L7K_B Chain B, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A],3L7K_C Chain C, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A],3L7K_D Chain D, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A],3L7L_A Chain A, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A],3L7L_B Chain B, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A],3L7L_C Chain C, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A],3L7L_D Chain D, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A] |
| 3L7M_A | 7.98e-22 | 36 | 385 | 350 | 707 | ChainA, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A],3L7M_B Chain B, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A],3L7M_C Chain C, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A],3L7M_D Chain D, Teichoic acid biosynthesis protein F [Staphylococcus epidermidis RP62A] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8RKI5 | 1.56e-29 | 34 | 393 | 27 | 393 | Teichoic acid poly(glycerol phosphate) polymerase OS=Bacillus spizizenii (strain ATCC 23059 / NRRL B-14472 / W23) OX=655816 GN=tarF PE=1 SV=1 |
| Q2G1C1 | 5.66e-27 | 33 | 392 | 23 | 387 | Teichoic acid glycerol-phosphate transferase OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=tarF PE=1 SV=1 |
| P13485 | 2.48e-24 | 33 | 368 | 376 | 712 | Teichoic acid poly(glycerol phosphate) polymerase OS=Bacillus subtilis (strain 168) OX=224308 GN=tagF PE=1 SV=1 |
| Q5HLM5 | 2.67e-22 | 36 | 385 | 350 | 707 | Teichoic acid poly(glycerol phosphate) polymerase OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=tagF PE=1 SV=1 |
| Q8RKJ1 | 2.75e-20 | 34 | 388 | 24 | 376 | Teichoic acid ribitol-phosphate primase OS=Bacillus spizizenii (strain ATCC 23059 / NRRL B-14472 / W23) OX=655816 GN=tarK PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
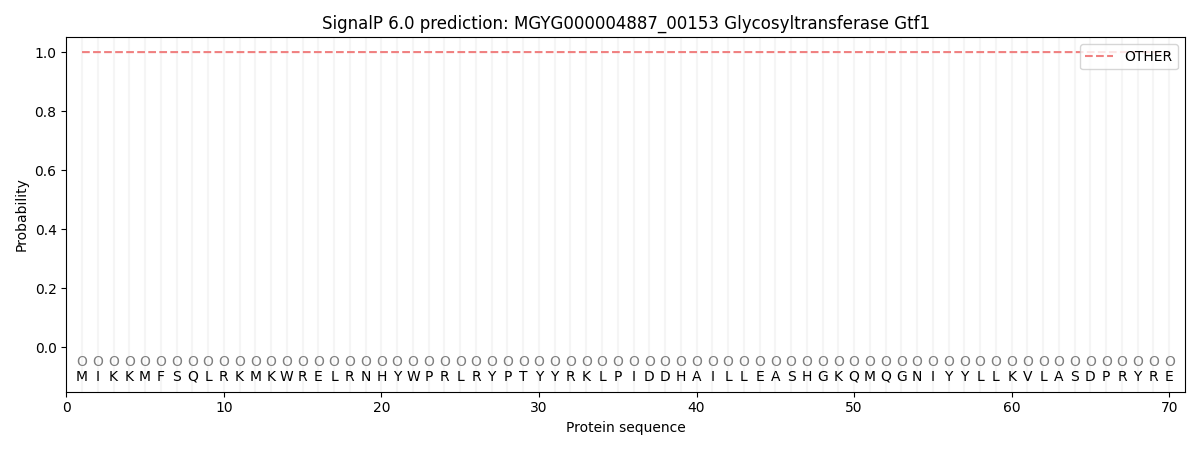
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000046 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
