You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000008_01753
You are here: Home > Sequence: MGYG000000008_01753
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
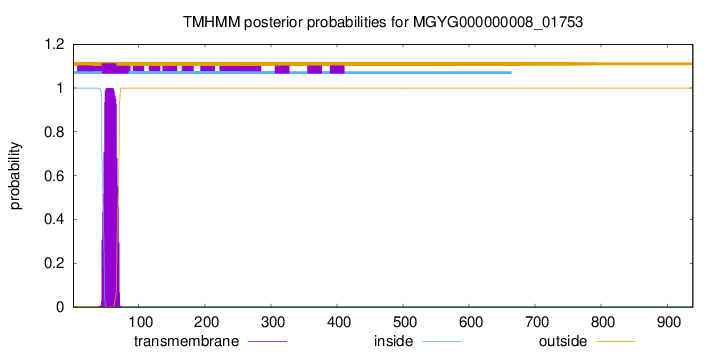
TMHMM annotations
Basic Information help
| Species | Lactobacillus johnsonii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Lactobacillus; Lactobacillus johnsonii | |||||||||||
| CAZyme ID | MGYG000000008_01753 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9014; End: 11833 Strand: + | |||||||||||
Full Sequence Download help
| MKNLKEKIRK FLTAGPEVKT LQSESDESQW KFYSGIVYLT LRRVFHYFIL IAFFGLFLLI | 60 |
| GFGGGYALGI VRNQPIPTIS ELNHQINHAQ NSATLYYAGN KKIATVRPDT VTKKASESEL | 120 |
| TPLVKNAVTA TEDENFYIHK GVLPKSLVRA VLSELTGVGV QTGGSTLTQQ LVKMQFLTSQ | 180 |
| TTWRRKVTEM FYAHKIEKHF SKEDILRAYL NAAPYGKNNR GENIVGIKTA AEGIFDKSIS | 240 |
| ELNLPQAAFI AGLPQSPSVY TPYRLNGKVK SDLDLAMRRK DIVLFRMYRN GDISKKAYLA | 300 |
| AKKYDLRADF LAPEKAPKQK KQSGYLYNLV MNKSASLLAE NLIEQDGLKV SDVKQDTNRY | 360 |
| NQYLTSATEL LHQKGYHIKT TIRKTLYQTM QQVVKQNRYG QDRTSRDFDS STNKWVNTTE | 420 |
| HVENGSVVID NATGKVLAFS GGVDFKNSQI NHAFDTYRSP GSSIKPYLVY GPAIEHKLIS | 480 |
| SQTAIADFPT RFGNYIPTDY NSTVENRFIS AQEALSKSYN LPAVNLYSKL VNDKNINLRQ | 540 |
| DMKKLGLNLS KSEFENLGLA LGGTDYGFSV ADNASAFSNF YNNGKRADPY YIEEIQDPSG | 600 |
| RVIYKHKQNP QKVFSAGTSY IMQKMLHQVV TKGTASSLTG TLKFDYKNLI GKTGTSNDYR | 660 |
| DIWFNGSTPG ITISSWMGYD NFYGHSYNLD SNSSETNLNL WANIANALYK EDPSIFKLND | 720 |
| APSRPSSVYK NSVLARTGTL PGTVSYDGDN IDLSGKKTTA LSLSPAPNAT ARFGIGGSTK | 780 |
| DYNLFYDYLE GKNNDYGKVL IYTGKTISKK KNINSLFAVA DGSTSENAKN YYGKNHEVYR | 840 |
| ESNNNSSSST NRNVGANDEQ ARSTGQGGNN NSSSTTRSTT STRESPSSSQ NNRSESSNAN | 900 |
| EANEVNSNIS PSESAPTSGS ASTEATAGDA AADNAPTAP | 939 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 111 | 288 | 2.7e-52 | 0.9322033898305084 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 3.48e-146 | 42 | 770 | 9 | 660 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| TIGR02074 | PBP_1a_fam | 3.25e-129 | 119 | 710 | 9 | 530 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG5009 | MrcA | 3.68e-91 | 47 | 683 | 7 | 695 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| COG4953 | PbpC | 1.94e-78 | 119 | 726 | 65 | 565 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 4.18e-65 | 118 | 288 | 19 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMT68502.1 | 0.0 | 1 | 939 | 1 | 939 |
| AYN49307.1 | 0.0 | 1 | 939 | 1 | 939 |
| AZZ67017.1 | 0.0 | 1 | 939 | 1 | 939 |
| AEB92818.1 | 0.0 | 1 | 939 | 1 | 939 |
| QLL67811.1 | 0.0 | 1 | 939 | 1 | 939 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JE5_A | 1.24e-135 | 61 | 791 | 3 | 717 | StructuralAnd Mechanistic Basis Of Penicillin Binding Protein Inhibition By Lactivicins [Streptococcus pneumoniae R6],2JE5_B Structural And Mechanistic Basis Of Penicillin Binding Protein Inhibition By Lactivicins [Streptococcus pneumoniae R6] |
| 2FFF_B | 1.93e-65 | 325 | 791 | 2 | 452 | OpenForm of a Class A Transpeptidase Domain [Streptococcus pneumoniae] |
| 2BG1_A | 2.83e-65 | 325 | 791 | 41 | 491 | Activesite restructuring regulates ligand recognition in classA Penicillin-binding proteins (PBPs) [Streptococcus pneumoniae R6],2XD5_A Structural insights into the catalytic mechanism and the role of Streptococcus pneumoniae PBP1b [Streptococcus pneumoniae R6],2XD5_B Structural insights into the catalytic mechanism and the role of Streptococcus pneumoniae PBP1b [Streptococcus pneumoniae R6] |
| 2XD1_A | 2.83e-65 | 325 | 791 | 41 | 491 | ACTIVESITE RESTRUCTURING REGULATES LIGAND RECOGNITION IN CLASS A PENICILLIN-BINDING PROTEINS [Streptococcus pneumoniae R6],2XD1_B ACTIVE SITE RESTRUCTURING REGULATES LIGAND RECOGNITION IN CLASS A PENICILLIN-BINDING PROTEINS [Streptococcus pneumoniae R6] |
| 2UWX_A | 1.39e-64 | 325 | 791 | 41 | 491 | Activesite restructuring regulates ligand recognition in class A penicillin-binding proteins [Streptococcus pneumoniae R6] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38050 | 1.91e-68 | 116 | 680 | 71 | 571 | Penicillin-binding protein 1F OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpF PE=2 SV=2 |
| A7GHV1 | 2.07e-57 | 124 | 746 | 88 | 676 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Langeland / NCTC 10281 / Type F) OX=441772 GN=pbpA PE=3 SV=1 |
| P40750 | 2.40e-57 | 124 | 709 | 88 | 624 | Penicillin-binding protein 4 OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpD PE=1 SV=2 |
| A5I6G4 | 3.91e-57 | 124 | 746 | 88 | 676 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Hall / ATCC 3502 / NCTC 13319 / Type A) OX=441771 GN=pbpA PE=3 SV=1 |
| A7FY32 | 3.91e-57 | 124 | 746 | 88 | 676 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain ATCC 19397 / Type A) OX=441770 GN=pbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
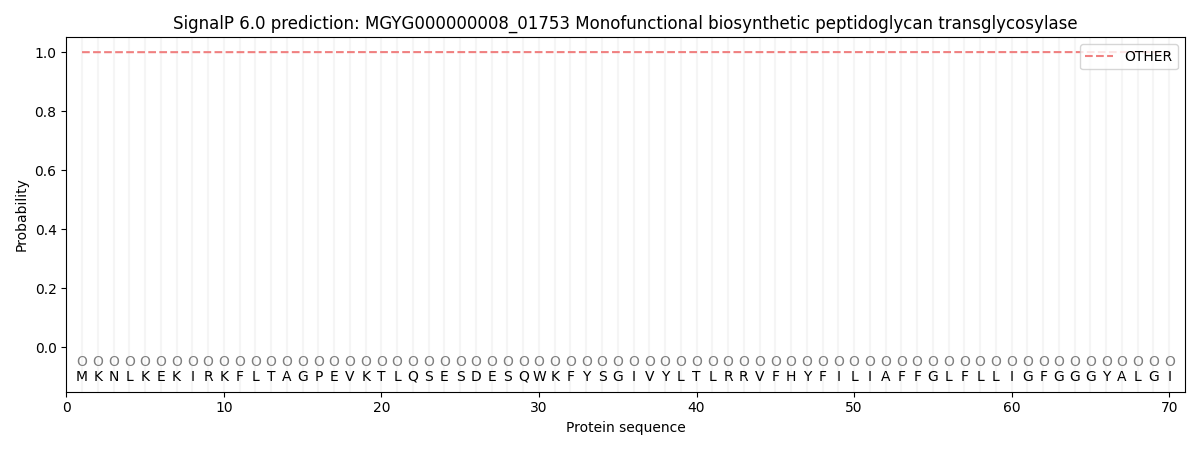
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999860 | 0.000139 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |